Risk Factors for Gut Dysbiosis in Early Life
Abstract
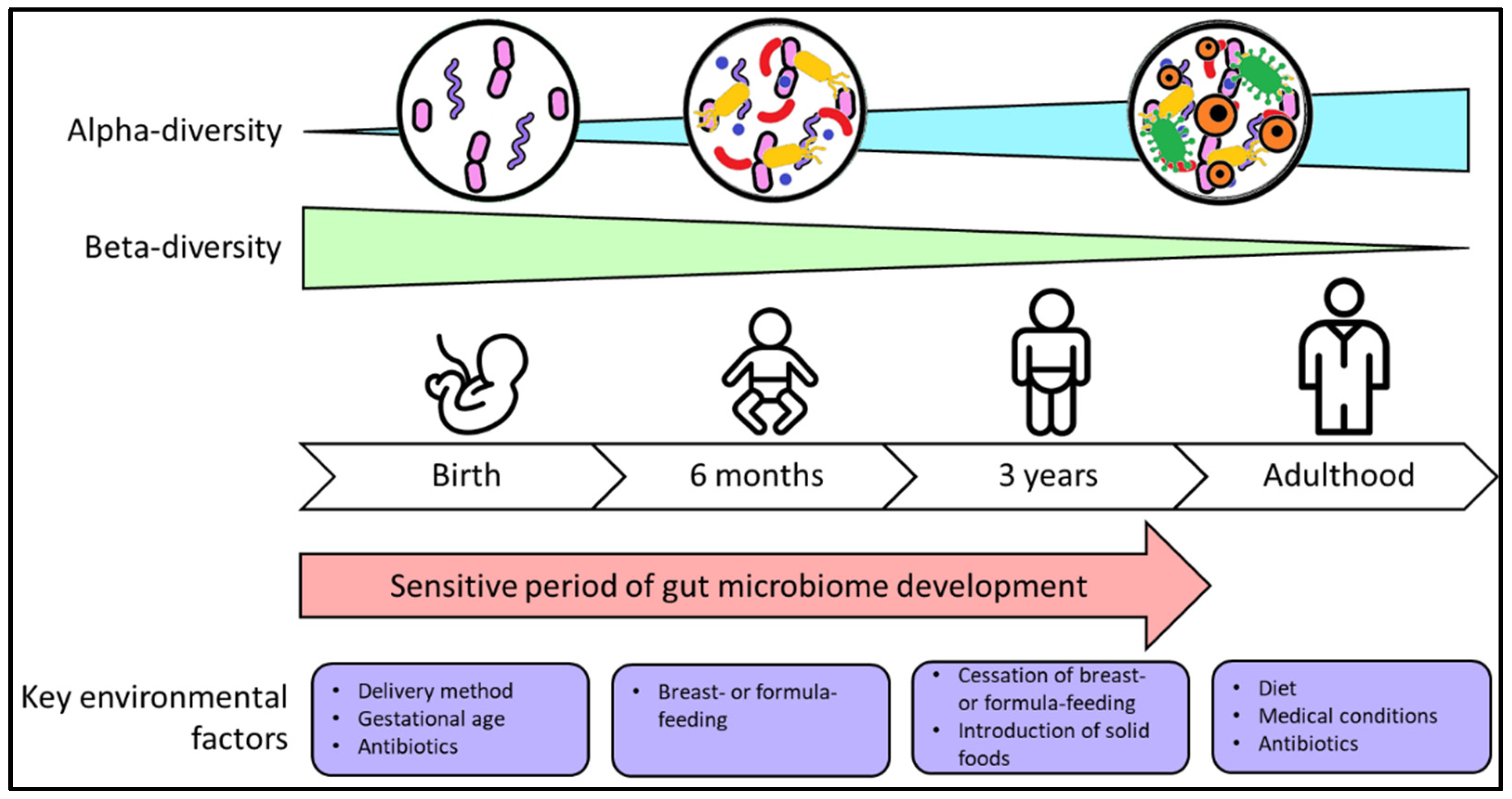
:1. Introduction
2. Factors That Shape Development of the Gut Microbiome in Infancy and Early Childhood
2.1. Parturition
2.2. Early Postnatal Life
2.3. Genetics
2.4. Infant Feeding
2.5. Introduction of Solid Foods
3. Environmental and Lifestyle Factors That Contribute to Gut Dysbiosis
3.1. Delivery Method
3.2. Antibiotic Use
3.3. Other Factors
4. Links between Gut Dysbiosis and Disease
4.1. GIT-Related Disorders
4.2. Central Nervous System-Related Disorders
4.3. Immunity
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Qin, J.; Li, R.; Raes, J.; Arumugam, M.; Burgdorf, K.S.; Manichanh, C.; Nielsen, T.; Pons, N.; Levenez, F.; Yamada, T.; et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 2010, 464, 59–65. [Google Scholar] [CrossRef] [Green Version]
- Hooper, L.V.; Gordon, J.I. Commensal host-bacterial relationships in the gut. Science 2001, 292, 1115–1118. [Google Scholar] [CrossRef]
- Laforest-Lapointe, I.; Arrieta, M.C. Patterns of early-life gut microbial colonization during human immune development: An ecological perspective. Front. Immunol. 2017, 8, 788. [Google Scholar] [CrossRef] [Green Version]
- Adamek, K.; Skonieczna-Zydecka, K.; Wegrzyn, D.; Loniewska, B. Prenatal and early childhood development of gut microbiota. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 9667–9680. [Google Scholar] [CrossRef]
- Wang, J.Z.; Du, W.T.; Xu, Y.L.; Cheng, S.Z.; Liu, Z.J. Gut microbiome-based medical methodologies for early-stage disease prevention. Microb. Pathog. 2017, 105, 122–130. [Google Scholar] [CrossRef]
- Tang, M.L.; Mullins, R.J. Food allergy: Is prevalence increasing? Intern. Med. J. 2017, 47, 256–261. [Google Scholar] [CrossRef]
- Dharmage, S.C.; Perret, J.L.; Custovic, A. Epidemiology of asthma in children and adults. Front. Pediatr. 2019, 7, 246. [Google Scholar] [CrossRef]
- Fang, X.; Henao-Mejia, J.; Henrickson, S.E. Obesity and immune status in children. Curr. Opin. Pediatr. 2020, 32, 805–815. [Google Scholar] [CrossRef] [PubMed]
- Cook, M.; Douglass, J.; Mallon, D.; Smith, J.; Wong, M.; Mullins, R. Economic impact of allergies. Access Econ. 2007. Available online: https://www.allergy.org.au/images/stories/pospapers/2007_economic_impact_allergies_report_13nov.pdf. (accessed on 25 September 2021).
- Renz, H.; Skevaki, C. Early life microbial exposures and allergy risks: Opportunities for prevention. Nat. Rev. Immunol. 2021, 21, 177–191. [Google Scholar] [CrossRef] [PubMed]
- Lambrecht, B.N.; Hammad, H. The immunology of the allergy epidemic and the hygiene hypothesis. Nat. Immunol. 2017, 18, 1076–1083. [Google Scholar] [CrossRef] [PubMed]
- De Filippo, C.; Cavalieri, D.; Di Paola, M.; Ramazzotti, M.; Poullet, J.B.; Massart, S.; Collini, S.; Pieraccini, G.; Lionetti, P. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc. Natl. Acad. Sci. USA 2010, 107, 14691–14696. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spor, A.; Koren, O.; Ley, R. Unravelling the effects of the environment and host genotype on the gut microbiome. Nat. Rev. Microbiol. 2011, 9, 279–290. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez, J.M.; Murphy, K.; Stanton, C.; Ross, R.P.; Kober, O.I.; Juge, N.; Avershina, E.; Rudi, K.; Narbad, A.; Jenmalm, M.C.; et al. The composition of the gut microbiota throughout life, with an emphasis on early life. Microb. Ecol. Health Dis. 2015, 26, 26050. [Google Scholar] [CrossRef]
- Yatsunenko, T.; Rey, F.E.; Manary, M.J.; Trehan, I.; Dominguez-Bello, M.G.; Contreras, M.; Magris, M.; Hidalgo, G.; Baldassano, R.N.; Anokhin, A.P.; et al. Human gut microbiome viewed across age and geography. Nature 2012, 486, 222–227. [Google Scholar] [CrossRef]
- Perez-Munoz, M.E.; Arrieta, M.C.; Ramer-Tait, A.E.; Walter, J. A critical assessment of the “sterile womb” and “in utero colonization” hypotheses: Implications for research on the pioneer infant microbiome. Microbiome 2017, 5, 48. [Google Scholar] [CrossRef]
- Mishra, A.; Lai, G.C.; Yao, L.J.; Aung, T.T.; Shental, N.; Rotter-Maskowitz, A.; Shepherdson, E.; Singh, G.S.N.; Pai, R.; Shanti, A.; et al. Microbial exposure during early human development primes fetal immune cells. Cell 2021, 184, 3394–3409.e3320. [Google Scholar] [CrossRef]
- Tanaka, M.; Nakayama, J. Development of the gut microbiota in infancy and its impact on health in later life. Allergol. Int. 2017, 66, 515–522. [Google Scholar] [CrossRef]
- Aagaard, K.; Ma, J.; Antony, K.M.; Ganu, R.; Petrosino, J.; Versalovic, J. The placenta harbors a unique microbiome. Sci. Transl. Med. 2014, 6, 237ra265. [Google Scholar] [CrossRef] [Green Version]
- Rautava, S.; Collado, M.C.; Salminen, S.; Isolauri, E. Probiotics modulate host-microbe interaction in the placenta and fetal gut: A randomized, double-blind, placebo-controlled trial. Neonatology 2012, 102, 178–184. [Google Scholar] [CrossRef]
- Jimenez, E.; Fernandez, L.; Marin, M.L.; Martin, R.; Odriozola, J.M.; Nueno-Palop, C.; Narbad, A.; Olivares, M.; Xaus, J.; Rodriguez, J.M. Isolation of commensal bacteria from umbilical cord blood of healthy neonates born by cesarean section. Curr. Microbiol. 2005, 51, 270–274. [Google Scholar] [CrossRef]
- Jimenez, E.; Marin, M.L.; Martin, R.; Odriozola, J.M.; Olivares, M.; Xaus, J.; Fernandez, L.; Rodriguez, J.M. Is meconium from healthy newborns actually sterile? Res. Microbiol. 2008, 159, 187–193. [Google Scholar] [CrossRef]
- Houghteling, P.D.; Walker, W.A. Why is initial bacterial colonization of the intestine important to infants’ and children’s health? J. Pediatr. Gastroenterol. Nutr. 2015, 60, 294–307. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dogra, S.; Sakwinska, O.; Soh, S.E.; Ngom-Bru, C.; Bruck, W.M.; Berger, B.; Brussow, H.; Karnani, N.; Lee, Y.S.; Yap, F.; et al. Rate of establishing the gut microbiota in infancy has consequences for future health. Gut Microbes 2015, 6, 321–325. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Backhed, F.; Roswall, J.; Peng, Y.; Feng, Q.; Jia, H.; Kovatcheva-Datchary, P.; Li, Y.; Xia, Y.; Xie, H.; Zhong, H.; et al. Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe 2015, 17, 852. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Almgren, M. Benefits of skin-to-skin contact during the neonatal period: Governed by epigenetic mechanisms? Genes Dis. 2018, 5, 24–26. [Google Scholar] [CrossRef] [PubMed]
- van den Elsen, L.W.J.; Garssen, J.; Burcelin, R.; Verhasselt, V. Shaping the gut microbiota by breastfeeding: The gateway to allergy prevention? Front. Pediatr. 2019, 7, 47. [Google Scholar] [CrossRef] [PubMed]
- Sela, D.A.; Chapman, J.; Adeuya, A.; Kim, J.H.; Chen, F.; Whitehead, T.R.; Lapidus, A.; Rokhsar, D.S.; Lebrilla, C.B.; German, J.B.; et al. The genome sequence of Bifidobacterium longum reveals adaptations for milk utilization within the infant microbiome. Proc. Natl. Acad. Sci. USA 2008, 105, 18964. [Google Scholar] [CrossRef] [Green Version]
- Bergström, A.; Skov, T.H.; Bahl, M.I.; Roager, H.M.; Christensen, L.B.; Ejlerskov, K.T.; Mølgaard, C.; Michaelsen, K.F.; Licht, T.R. Establishment of intestinal microbiota during early life: A longitudinal, explorative study of a large cohort of Danish infants. Appl. Environ. Microbiol. 2014, 80, 2889–2900. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Faith, J.J.; Guruge, J.L.; Charbonneau, M.; Subramanian, S.; Seedorf, H.; Goodman, A.L.; Clemente, J.C.; Knight, R.; Heath, A.C.; Leibel, R.L.; et al. The long-term stability of the human gut microbiota. Science 2013, 341, 1237439. [Google Scholar] [CrossRef] [Green Version]
- Rajilić-Stojanović, M.; Heilig, H.G.; Tims, S.; Zoetendal, E.G.; de Vos, W.M. Long-term monitoring of the human intestinal microbiota composition. Environ. Microbiol. 2012. [Google Scholar] [CrossRef]
- Yassour, M.; Vatanen, T.; Siljander, H.; Hämäläinen, A.M.; Härkönen, T.; Ryhänen, S.J.; Franzosa, E.A.; Vlamakis, H.; Huttenhower, C.; Gevers, D.; et al. Natural history of the infant gut microbiome and impact of antibiotic treatment on bacterial strain diversity and stability. Sci. Transl. Med. 2016, 8, 343ra381. [Google Scholar] [CrossRef] [Green Version]
- Koenig, J.E.; Spor, A.; Scalfone, N.; Fricker, A.D.; Stombaugh, J.; Knight, R.; Angenent, L.T.; Ley, R.E. Succession of microbial consortia in the developing infant gut microbiome. Proc. Natl. Acad. Sci. USA 2011, 108 (Suppl. S1), 4578–4585. [Google Scholar] [CrossRef] [Green Version]
- Derrien, M.; Alvarez, A.-S.; de Vos, W.M. The gut microbiota in the first decade of life. Trends Microbiol. 2019, 27, 997–1010. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhong, H.; Penders, J.; Shi, Z.; Ren, H.; Cai, K.; Fang, C.; Ding, Q.; Thijs, C.; Blaak, E.E.; Stehouwer, C.D.A.; et al. Impact of early events and lifestyle on the gut microbiota and metabolic phenotypes in young school-age children. Microbiome 2019, 7, 2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deering, K.E.; Devine, A.; O’Sullivan, T.A.; Lo, J.; Boyce, M.C.; Christophersen, C.T. Characterizing the Composition of the Pediatric Gut Microbiome: A Systematic Review. Nutrients 2020, 12, 16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Thingholm, L.B.; Skiecevičienė, J.; Rausch, P.; Kummen, M.; Hov, J.R.; Degenhardt, F.; Heinsen, F.-A.; Rühlemann, M.C.; Szymczak, S.; et al. Genome-wide association analysis identifies variation in vitamin D receptor and other host factors influencing the gut microbiota. Nat. Genet. 2016, 48, 1396–1406. [Google Scholar] [CrossRef] [PubMed]
- Bonder, M.J.; Kurilshikov, A.; Tigchelaar, E.F.; Mujagic, Z.; Imhann, F.; Vila, A.V.; Deelen, P.; Vatanen, T.; Schirmer, M.; Smeekens, S.P.; et al. The effect of host genetics on the gut microbiome. Nat. Genet. 2016, 48, 1407–1412. [Google Scholar] [CrossRef]
- Blekhman, R.; Goodrich, J.K.; Huang, K.; Sun, Q.; Bukowski, R.; Bell, J.T.; Spector, T.D.; Keinan, A.; Ley, R.E.; Gevers, D.; et al. Host genetic variation impacts microbiome composition across human body sites. Genome Biol. 2015, 16, 191. [Google Scholar] [CrossRef] [Green Version]
- Xu, F.; Fu, Y.; Sun, T.-Y.; Jiang, Z.; Miao, Z.; Shuai, M.; Gou, W.; Ling, C.-W.; Yang, J.; Wang, J.; et al. The interplay between host genetics and the gut microbiome reveals common and distinct microbiome features for complex human diseases. Microbiome 2020, 8, 145. [Google Scholar] [CrossRef]
- Turpin, W.; Espin-Garcia, O.; Xu, W.; Silverberg, M.S.; Kevans, D.; Smith, M.I.; Guttman, D.S.; Griffiths, A.; Panaccione, R.; Otley, A.; et al. Association of host genome with intestinal microbial composition in a large healthy cohort. Nat. Genet. 2016, 48, 1413–1417. [Google Scholar] [CrossRef]
- Goodrich, J.K.; Waters, J.L.; Poole, A.C.; Sutter, J.L.; Koren, O.; Blekhman, R.; Beaumont, M.; Van Treuren, W.; Knight, R.; Bell, J.T.; et al. Human genetics shape the gut microbiome. Cell 2014, 159, 789–799. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- The Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature 2012, 486, 207–214. [Google Scholar] [CrossRef] [Green Version]
- Deschasaux, M.; Bouter, K.E.; Prodan, A.; Levin, E.; Groen, A.K.; Herrema, H.; Tremaroli, V.; Bakker, G.J.; Attaye, I.; Pinto-Sietsma, S.-J.; et al. Depicting the composition of gut microbiota in a population with varied ethnic origins but shared geography. Nat. Med. 2018, 24, 1526–1531. [Google Scholar] [CrossRef]
- Gaulke, C.A.; Sharpton, T.J. The influence of ethnicity and geography on human gut microbiome composition. Nat. Med. 2018, 24, 1495–1496. [Google Scholar] [CrossRef]
- Wang, M.; Li, M.; Wu, S.; Lebrilla, C.B.; Chapkin, R.S.; Ivanov, I.; Donovan, S.M. Fecal microbiota composition of breast-fed infants is correlated with human milk oligosaccharides consumed. J. Pediatr. Gastroenterol. Nutr. 2015, 60, 825–833. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iyengar, S.R.; Walker, W.A. Immune factors in breast milk and the development of atopic disease. J. Pediatr. Gastroenterol. Nutr. 2012, 55, 641–647. [Google Scholar] [CrossRef]
- Dzidic, M.; Mira, A.; Artacho, A.; Abrahamsson, T.R.; Jenmalm, M.C.; Collado, M.C. Allergy development is associated with consumption of breastmilk with a reduced microbial richness in the first month of life. Pediatr. Allergy Immunol. 2019, 31, 250–257. [Google Scholar] [CrossRef] [PubMed]
- Cacho, N.T.; Lawrence, R.M. Innate immunity and breast milk. Front. Immunol. 2017, 8, 584. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reguigne-Arnould, I.; Couillin, P.; Mollicone, R.; Faure, S.; Fletcher, A.; Kelly, R.J.; Lowe, J.B.; Oriol, R. Relative positions of two clusters of human alpha-L-fucosyltransferases in 19q (FUT1-FUT2) and 19p (FUT6-FUT3-FUT5) within the microsatellite genetic map of chromosome 19. Cytogenet. Cell Genet. 1995, 71, 158–162. [Google Scholar] [CrossRef]
- Hao, H.; Zhu, L.; Faden, H.S. The milk-based diet of infancy and the gut microbiome. Gastroenterol. Rep. 2019, 7, 246–249. [Google Scholar] [CrossRef]
- Turpin, W.; Bedrani, L.; Espin-Garcia, O.; Xu, W.; Silverberg, M.S.; Smith, M.I.; Guttman, D.S.; Griffiths, A.; Moayyedi, P.; Panaccione, R.; et al. FUT2 genotype and secretory status are not associated with fecal microbial composition and inferred function in healthy subjects. Gut Microbes 2018, 9, 357–368. [Google Scholar] [CrossRef]
- Palmer, C.; Bik, E.M.; DiGiulio, D.B.; Relman, D.A.; Brown, P.O. Development of the human infant intestinal microbiota. PLoS Biol. 2007, 5, e177. [Google Scholar] [CrossRef] [Green Version]
- Differding, M.K.; Benjamin-Neelon, S.E.; Hoyo, C.; Østbye, T.; Mueller, N.T. Timing of complementary feeding is associated with gut microbiota diversity and composition and short chain fatty acid concentrations over the first year of life. BMC Microbiol. 2020, 20, 56. [Google Scholar] [CrossRef] [Green Version]
- Rutayisire, E.; Huang, K.; Liu, Y.; Tao, F. The mode of delivery affects the diversity and colonization pattern of the gut microbiota during the first year of infants’ life: A systematic review. BMC Gastroenterol. 2016, 16, 86. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sevelsted, A.; Stokholm, J.; Bønnelykke, K.; Bisgaard, H. Cesarean section and chronic immune disorders. Pediatrics 2015, 135, e92–e98. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dominguez-Bello, M.G.; De Jesus-Laboy, K.M.; Shen, N.; Cox, L.M.; Amir, A.; Gonzalez, A.; Bokulich, N.A.; Song, S.J.; Hoashi, M.; Rivera-Vinas, J.I.; et al. Partial restoration of the microbiota of cesarean-born infants via vaginal microbial transfer. Nat. Med. 2016, 22, 250–253. [Google Scholar] [CrossRef]
- Betrán, A.P.; Ye, J.; Moller, A.B.; Zhang, J.; Gülmezoglu, A.M.; Torloni, M.R. The increasing trend in caesarean section rates: Global, regional and national estimates: 1990–2014. PLoS ONE 2016, 11, e0148343. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dominguez-Bello, M.G.; Costello, E.K.; Contreras, M.; Magris, M.; Hidalgo, G.; Fierer, N.; Knight, R. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc. Natl. Acad. Sci. USA 2010, 107, 11971–11975. [Google Scholar] [CrossRef] [Green Version]
- Levin, A.M.; Sitarik, A.R.; Havstad, S.L.; Fujimura, K.E.; Wegienka, G.; Cassidy-Bushrow, A.E.; Kim, H.; Zoratti, E.M.; Lukacs, N.W.; Boushey, H.A.; et al. Joint effects of pregnancy, sociocultural, and environmental factors on early life gut microbiome structure and diversity. Sci. Rep. 2016, 6, 31775. [Google Scholar] [CrossRef] [PubMed]
- Dogra, S.; Sakwinska, O.; Soh, S.E.; Ngom-Bru, C.; Brück, W.M.; Berger, B.; Brüssow, H.; Lee, Y.S.; Yap, F.; Chong, Y.S.; et al. Dynamics of infant gut microbiota are influenced by delivery mode and gestational duration and are associated with subsequent adiposity. mBio 2015, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hesla, H.M.; Stenius, F.; Jäderlund, L.; Nelson, R.; Engstrand, L.; Alm, J.; Dicksved, J. Impact of lifestyle on the gut microbiota of healthy infants and their mothers—the ALADDIN birth cohort. FEMS Microbiol. Ecol. 2014, 90, 791–801. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Renz-Polster, H.; David, M.R.; Buist, A.S.; Vollmer, W.M.; O’Connor, E.A.; Frazier, E.A.; Wall, M.A. Caesarean section delivery and the risk of allergic disorders in childhood. Clin. Exp. Allergy 2005, 35, 1466–1472. [Google Scholar] [CrossRef]
- Imoto, N.; Morita, H.; Amanuma, F.; Maruyama, H.; Watanabe, S.; Hashiguchi, N. Maternal antimicrobial use at delivery has a stronger impact than mode of delivery on bifidobacterial colonization in infants: A pilot study. J. Perinatol. 2018, 38, 1174–1181. [Google Scholar] [CrossRef] [Green Version]
- Kingsbury, M.A.; Bilbo, S.D. The inflammatory event of birth: How oxytocin signaling may guide the development of the brain and gastrointestinal system. Front. Neuroendocrinol. 2019, 100794. [Google Scholar] [CrossRef]
- Strzepa, A.; Lobo, F.M.; Majewska-Szczepanik, M.; Szczepanik, M. Antibiotics and autoimmune and allergy diseases: Causative factor or treatment? Int. Immunopharmacol. 2018, 65, 328–341. [Google Scholar] [CrossRef]
- Metsala, J.; Lundqvist, A.; Virta, L.J.; Kaila, M.; Gissler, M.; Virtanen, S.M. Prenatal and post-natal exposure to antibiotics and risk of asthma in childhood. Clin. Exp. Allergy 2015, 45, 137–145. [Google Scholar] [CrossRef]
- van der Waaij, D.; Nord, C.E. Development and persistence of multi-resistance to antibiotics in bacteria; an analysis and a new approach to this urgent problem. Int. J. Antimicrob. Agents 2000, 16, 191–197. [Google Scholar] [CrossRef]
- Belizário, J.E.; Faintuch, J. Microbiome and gut dysbiosis. Exp. Suppl. 2018, 109, 459–476. [Google Scholar] [CrossRef] [PubMed]
- Zimmermann, P.; Curtis, N. The effect of antibiotics on the composition of the intestinal microbiota—A systematic review. J. Infect. 2019, 79, 471–489. [Google Scholar] [CrossRef] [PubMed]
- Hagan, T.; Cortese, M.; Rouphael, N.; Boudreau, C.; Linde, C.; Maddur, M.S.; Das, J.; Wang, H.; Guthmiller, J.; Zheng, N.Y.; et al. Antibiotics-driven gut microbiome perturbation alters immunity to vaccines in humans. Cell 2019, 178, 1313–1328.e1313. [Google Scholar] [CrossRef]
- Palleja, A.; Mikkelsen, K.H.; Forslund, S.K.; Kashani, A.; Allin, K.H.; Nielsen, T.; Hansen, T.H.; Liang, S.; Feng, Q.; Zhang, C.; et al. Recovery of gut microbiota of healthy adults following antibiotic exposure. Nat. Microbiol. 2018, 3, 1255–1265. [Google Scholar] [CrossRef]
- Korpela, K.; Salonen, A.; Saxen, H.; Nikkonen, A.; Peltola, V.; Jaakkola, T.; de Vos, W.; Kolho, K.L. Antibiotics in early life associate with specific gut microbiota signatures in a prospective longitudinal infant cohort. Pediatr. Res. 2020, 88, 438–443. [Google Scholar] [CrossRef]
- Sun, L.; Zhang, X.; Zhang, Y.; Zheng, K.; Xiang, Q.; Chen, N.; Chen, Z.; Zhang, N.; Zhu, J.; He, Q. Antibiotic-Induced Disruption of Gut Microbiota Alters Local Metabolomes and Immune Responses. Front. Cell. Infect. Microbiol. 2019, 9, 99. [Google Scholar] [CrossRef] [PubMed]
- Ownby, D.R.; Johnson, C.C.; Peterson, E.L. Exposure to dogs and cats in the first year of life and risk of allergic sensitization at 6 to 7 years of age. JAMA 2002, 288, 963–972. [Google Scholar] [CrossRef] [Green Version]
- Azad, M.B.; Konya, T.; Maughan, H.; Guttman, D.S.; Field, C.J.; Sears, M.R.; Becker, A.B.; Scott, J.A.; Kozyrskyj, A.L. Infant gut microbiota and the hygiene hypothesis of allergic disease: Impact of household pets and siblings on microbiota composition and diversity. Allergy Asthma Clin. Immunol. 2013, 9, 15. [Google Scholar] [CrossRef] [Green Version]
- Nermes, M.; Endo, A.; Aarnio, J.; Salminen, S.; Isolauri, E. Furry pets modulate gut microbiota composition in infants at risk for allergic disease. J. Allergy Clin. Immunol. 2015, 136, 1688–1690.e1681. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tun, H.M.; Konya, T.; Takaro, T.K.; Brook, J.R.; Chari, R.; Field, C.J.; Guttman, D.S.; Becker, A.B.; Mandhane, P.J.; Turvey, S.E.; et al. Exposure to household furry pets influences the gut microbiota of infant at 3-4 months following various birth scenarios. Microbiome 2017, 5, 40. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Collado, M.C.; Cernada, M.; Neu, J.; Pérez-Martínez, G.; Gormaz, M.; Vento, M. Factors influencing gastrointestinal tract and microbiota immune interaction in preterm infants. Pediatr. Res. 2015, 77, 726–731. [Google Scholar] [CrossRef]
- Arboleya, S.; Binetti, A.; Salazar, N.; Fernández, N.; Solís, G.; Hernández-Barranco, A.; Margolles, A.; de Los Reyes-Gavilán, C.G.; Gueimonde, M. Establishment and development of intestinal microbiota in preterm neonates. FEMS Microbiol. Ecol. 2012, 79, 763–772. [Google Scholar] [CrossRef] [Green Version]
- Young, G.R.; van der Gast, C.J.; Smith, D.L.; Berrington, J.E.; Embleton, N.D.; Lanyon, C. Acquisition and development of the extremely preterm infant microbiota across multiple anatomical sites. J. Pediatr. Gastroenterol. Nutr. 2020, 70, 12–19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morais, J.; Marques, C.; Teixeira, D.; Durão, C.; Faria, A.; Brito, S.; Cardoso, M.; Macedo, I.; Pereira, E.; Tomé, T.; et al. Extremely preterm neonates have more Lactobacillus in meconium than very preterm neonates-the in utero microbial colonization hypothesis. Gut Microbes 2020, 12, 1785804. [Google Scholar] [CrossRef] [PubMed]
- Henderickx, J.G.E.; Zwittink, R.D.; van Lingen, R.A.; Knol, J.; Belzer, C. The preterm gut microbiota: An inconspicuous challenge in nutritional neonatal care. Front. Cell Infect. Microbiol. 2019, 9, 85. [Google Scholar] [CrossRef]
- Tauchi, H.; Yahagi, K.; Yamauchi, T.; Hara, T.; Yamaoka, R.; Tsukuda, N.; Watanabe, Y.; Tajima, S.; Ochi, F.; Iwata, H.; et al. Gut microbiota development of preterm infants hospitalised in intensive care units. Benef. Microbes 2019, 10, 641–651. [Google Scholar] [CrossRef]
- Groer, M.W.; Miller, E.M.; D’Agata, A.; Ho, T.T.B.; Dutra, S.V.; Yoo, J.Y.; Yee, A.L.; Gilbert, J.A.; Dishaw, L.J. Contributors to dysbiosis in very-low-birth-weight infants. J. Obstet. Gynecol. Neonatal Nurs. 2020, 49, 232–242. [Google Scholar] [CrossRef]
- Carding, S.; Verbeke, K.; Vipond, D.T.; Corfe, B.M.; Owen, L.J. Dysbiosis of the gut microbiota in disease. Microb. Ecol. Health Dis. 2015, 26, 26191. [Google Scholar] [CrossRef]
- Frank, D.N.; St Amand, A.L.; Feldman, R.A.; Boedeker, E.C.; Harpaz, N.; Pace, N.R. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc. Natl. Acad. Sci. USA 2007, 104, 13780–13785. [Google Scholar] [CrossRef] [Green Version]
- Baumgart, D.C.; Carding, S.R. Inflammatory bowel disease: Cause and immunobiology. Lancet 2007, 369, 1627–1640. [Google Scholar] [CrossRef]
- Hansen, J.; Gulati, A.; Sartor, R.B. The role of mucosal immunity and host genetics in defining intestinal commensal bacteria. Curr. Opin. Gastroenterol. 2010, 26, 564–571. [Google Scholar] [CrossRef]
- Krogius-Kurikka, L.; Lyra, A.; Malinen, E.; Aarnikunnas, J.; Tuimala, J.; Paulin, L.; Mäkivuokko, H.; Kajander, K.; Palva, A. Microbial community analysis reveals high level phylogenetic alterations in the overall gastrointestinal microbiota of diarrhoea-predominant irritable bowel syndrome sufferers. BMC Gastroenterol. 2009, 9, 95. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sudo, N. Microbiome, HPA axis and production of endocrine hormones in the gut. Adv. Exp. Med. Biol. 2014, 817, 177–194. [Google Scholar] [CrossRef] [PubMed]
- Collins, J.; Borojevic, R.; Verdu, E.F.; Huizinga, J.D.; Ratcliffe, E.M. Intestinal microbiota influence the early postnatal development of the enteric nervous system. Neurogastroenterol. Motil. 2014, 26, 98–107. [Google Scholar] [CrossRef] [PubMed]
- Bravo, J.A.; Forsythe, P.; Chew, M.V.; Escaravage, E.; Savignac, H.M.; Dinan, T.G.; Bienenstock, J.; Cryan, J.F. Ingestion of Lactobacillus strain regulates emotional behavior and central GABA receptor expression in a mouse via the vagus nerve. Proc. Natl. Acad. Sci. USA 2011, 108, 16050–16055. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Erny, D.; Hrabě de Angelis, A.L.; Jaitin, D.; Wieghofer, P.; Staszewski, O.; David, E.; Keren-Shaul, H.; Mahlakoiv, T.; Jakobshagen, K.; Buch, T.; et al. Host microbiota constantly control maturation and function of microglia in the CNS. Nat. Neurosci. 2015, 18, 965–977. [Google Scholar] [CrossRef]
- Silva, Y.P.; Bernardi, A.; Frozza, R.L. The Role of Short-Chain Fatty Acids From Gut Microbiota in Gut-Brain Communication. Front. Endocrinol. 2020, 11, 25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dalile, B.; Van Oudenhove, L.; Vervliet, B.; Verbeke, K. The role of short-chain fatty acids in microbiota–gut–brain communication. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 461–478. [Google Scholar] [CrossRef]
- Vuong, H.E.; Pronovost, G.N.; Williams, D.W.; Coley, E.J.L.; Siegler, E.L.; Qiu, A.; Kazantsev, M.; Wilson, C.J.; Rendon, T.; Hsiao, E.Y. The maternal microbiome modulates fetal neurodevelopment in mice. Nature 2020, 586, 281–286. [Google Scholar] [CrossRef]
- Sun, J.; Xu, J.; Ling, Y.; Wang, F.; Gong, T.; Yang, C.; Ye, S.; Ye, K.; Wei, D.; Song, Z.; et al. Fecal microbiota transplantation alleviated Alzheimer’s disease-like pathogenesis in APP/PS1 transgenic mice. Transl. Psychiatry 2019, 9, 189. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Finegold, S.M.; Dowd, S.E.; Gontcharova, V.; Liu, C.; Henley, K.E.; Wolcott, R.D.; Youn, E.; Summanen, P.H.; Granpeesheh, D.; Dixon, D.; et al. Pyrosequencing study of fecal microflora of autistic and control children. Anaerobe 2010, 16, 444–453. [Google Scholar] [CrossRef]
- Wang, L.; Christophersen, C.T.; Sorich, M.J.; Gerber, J.P.; Angley, M.T.; Conlon, M.A. Low relative abundances of the mucolytic bacterium Akkermansia muciniphila and Bifidobacterium spp. in feces of children with autism. Appl. Environ. Microbiol. 2011, 77, 6718–6721. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Christophersen, C.T.; Sorich, M.J.; Gerber, J.P.; Angley, M.T.; Conlon, M.A. Elevated fecal short chain fatty acid and ammonia concentrations in children with autism spectrum disorder. Dig. Dis. Sci. 2012, 57, 2096–2102. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Christophersen, C.T.; Sorich, M.J.; Gerber, J.P.; Angley, M.T.; Conlon, M.A. Increased abundance of Sutterella spp. and Ruminococcus torques in feces of children with autism spectrum disorder. Mol. Autism 2013, 4, 42. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sudo, N.; Sawamura, S.; Tanaka, K.; Aiba, Y.; Kubo, C.; Koga, Y. The requirement of intestinal bacterial flora for the development of an IgE production system fully susceptible to oral tolerance induction. J. Immunol. 1997, 159, 1739–1745. [Google Scholar]
- Bisgaard, H.; Li, N.; Bonnelykke, K.; Chawes, B.L.; Skov, T.; Paludan-Müller, G.; Stokholm, J.; Smith, B.; Krogfelt, K.A. Reduced diversity of the intestinal microbiota during infancy is associated with increased risk of allergic disease at school age. J. Allergy Clin. Immunol. 2011, 128, 646–652.e641–e645. [Google Scholar] [CrossRef]
- Fujimura, K.E.; Sitarik, A.R.; Havstad, S.; Lin, D.L.; Levan, S.; Fadrosh, D.; Panzer, A.R.; LaMere, B.; Rackaityte, E.; Lukacs, N.W.; et al. Neonatal gut microbiota associates with childhood multisensitized atopy and T cell differentiation. Nat. Med. 2016, 22, 1187–1191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ling, Z.; Li, Z.; Liu, X.; Cheng, Y.; Luo, Y.; Tong, X.; Yuan, L.; Wang, Y.; Sun, J.; Li, L.; et al. Altered fecal microbiota composition associated with food allergy in infants. Appl. Environ. Microbiol. 2014, 80, 2546–2554. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Azad, M.B.; Konya, T.; Guttman, D.S.; Field, C.J.; Sears, M.R.; HayGlass, K.T.; Mandhane, P.J.; Turvey, S.E.; Subbarao, P.; Becker, A.B.; et al. Infant gut microbiota and food sensitization: Associations in the first year of life. Clin. Exp. Allergy 2015, 45, 632–643. [Google Scholar] [CrossRef] [PubMed]
- Penders, J.; Thijs, C.; van den Brandt, P.A.; Kummeling, I.; Snijders, B.; Stelma, F.; Adams, H.; van Ree, R.; Stobberingh, E.E. Gut microbiota composition and development of atopic manifestations in infancy: The KOALA Birth Cohort Study. Gut 2007, 56, 661. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van Nimwegen, F.A.; Penders, J.; Stobberingh, E.E.; Postma, D.S.; Koppelman, G.H.; Kerkhof, M.; Reijmerink, N.E.; Dompeling, E.; van den Brandt, P.A.; Ferreira, I.; et al. Mode and place of delivery, gastrointestinal microbiota, and their influence on asthma and atopy. J. Allergy Clin. Immunol. 2011, 128, 948–955.e1–e3. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Parkin, K.; Christophersen, C.T.; Verhasselt, V.; Cooper, M.N.; Martino, D. Risk Factors for Gut Dysbiosis in Early Life. Microorganisms 2021, 9, 2066. https://doi.org/10.3390/microorganisms9102066
Parkin K, Christophersen CT, Verhasselt V, Cooper MN, Martino D. Risk Factors for Gut Dysbiosis in Early Life. Microorganisms. 2021; 9(10):2066. https://doi.org/10.3390/microorganisms9102066
Chicago/Turabian StyleParkin, Kimberley, Claus T. Christophersen, Valerie Verhasselt, Matthew N. Cooper, and David Martino. 2021. "Risk Factors for Gut Dysbiosis in Early Life" Microorganisms 9, no. 10: 2066. https://doi.org/10.3390/microorganisms9102066
APA StyleParkin, K., Christophersen, C. T., Verhasselt, V., Cooper, M. N., & Martino, D. (2021). Risk Factors for Gut Dysbiosis in Early Life. Microorganisms, 9(10), 2066. https://doi.org/10.3390/microorganisms9102066






