PCR and Culture Analysis of Streptococcus pneumoniae Nasopharyngeal Carriage in Healthy Children
Abstract
:1. Introduction
2. Materials and Methods
2.1. Original Surveys to Collect Samples and Identify S. pneumoniae by Culture and Quellung Serotyping
2.2. Identification of S. pneumoniae and Serotypes by qPCR
2.3. Statistical Analysis
3. Results
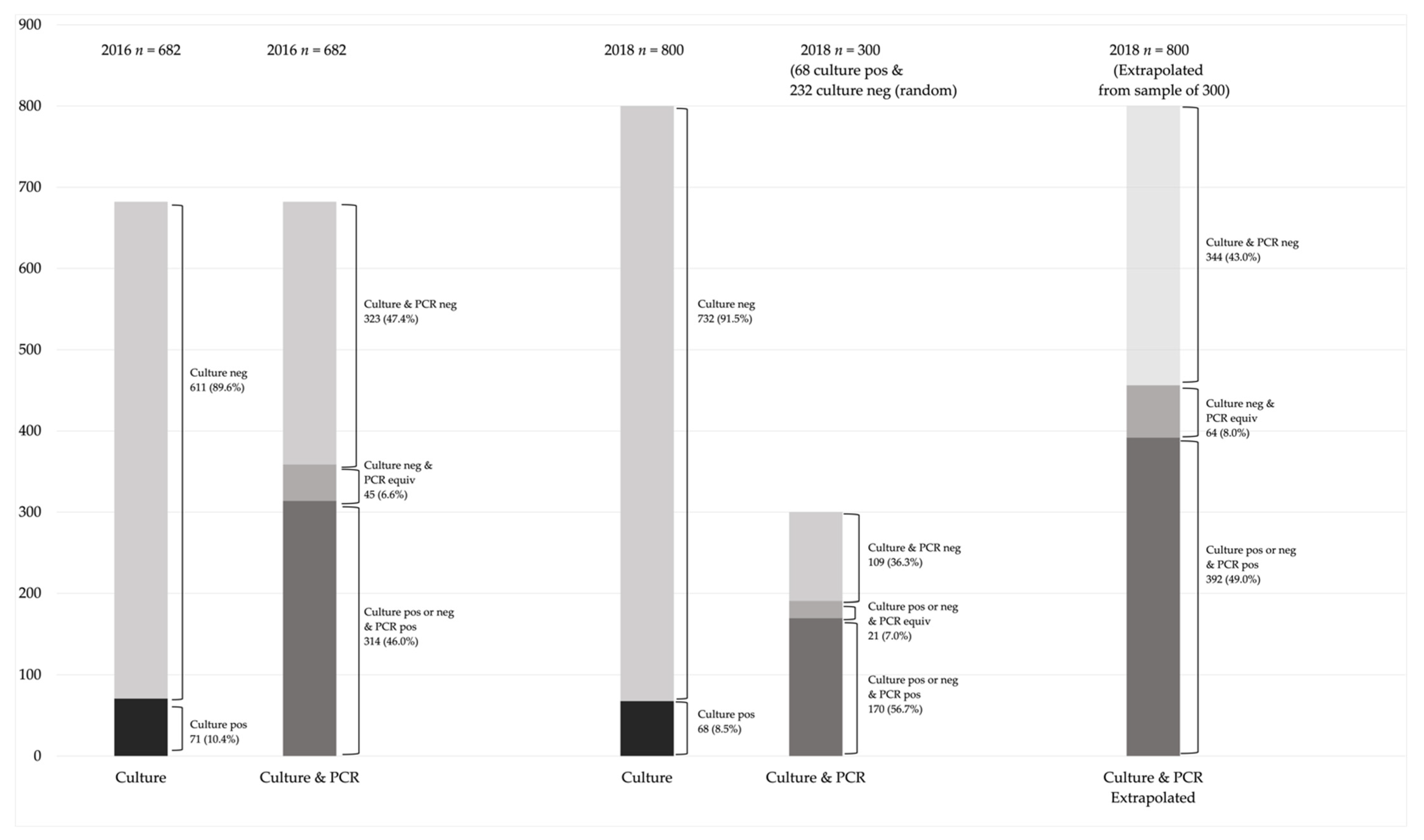
3.1. Analysis of 2016 Samples
3.2. Analysis of 2018 Samples
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bogaert, D.; de Groot, R.; Hermans, P.W.M. Streptococcus pneumoniae colonisation: The key to pneumococcal disease. Lancet Infect. Dis. 2004, 4, 144–154. [Google Scholar] [CrossRef]
- Gray, B.M.; Converse, G.M., 3rd; Dillon, H.C., Jr. Epidemiologic studies of Streptococcus pneumoniae in infants: Acquisition, carriage, and infection during the first 24 months of life. J. Infect. Dis. 1980, 142, 923–933. [Google Scholar] [CrossRef]
- Public Health Agency of Canada. Update on the Use of Pneumococcal Vaccines in Adults 65 Years of Age and Older—A Public Health Perspective: An Advisory Committee Statement (ACS) National Advisory Committee on Immunization (NACI). Available online: https://www.canada.ca/en/public-health/services/publications/healthy-living/update-on-the-use-of-pneumococcal-vaccines-in-adult.html (accessed on 13 March 2021).
- LeBlanc, J.J.; ElSherif, M.; Ye, L.; MacKinnon-Cameron, D.; Ambrose, A.; Hatchette, T.F.; Lang, A.L.S.; Gillis, H.D.; Martin, I.; Demczuk, W.; et al. Streptococcus pneumoniae serotype 3 is masking PCV13-mediated herd immunity in Canadian adults hospitalized with community acquired pneumonia: A study from the Serious Outcomes Surveillance (SOS) Network of the Canadian immunization research Network (CIRN). Vaccine 2019, 37, 5466–5473. [Google Scholar] [CrossRef]
- Ricketson, L.J.; Wood, M.L.; Vanderkooi, O.G.; MacDonald, J.C.; Martin, I.E.; Demczuk, W.H.; Kellner, J.D. Trends in asymptomatic nasopharyngeal colonization with S. pneumoniae after introduction of the 13-valent pneumococcal conjugate vaccine in Calgary, Canada. Pediatr. Infect. Dis. J. 2014, 33, 724–730. [Google Scholar] [CrossRef] [PubMed]
- Austrian, R. The quellung reaction, a neglected microbiologic technique. Mt. Sinai J. Med. N. Y. 1976, 43, 699–709. [Google Scholar]
- Da Gloria Carvalho, M.; Pimenta, F.C.; Jackson, D.; Roundtree, A.; Ahmad, Y.; Millar, E.V.; O’Brien, K.L.; Whitney, C.G.; Cohen, A.L.; Beall, B.W. Revisiting pneumococcal carriage by use of broth enrichment and PCR techniques for enhanced detection of carriage and serotypes. J. Clin. Microbiol. 2010, 48, 1611–1618. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hamer, D.H.; Egas, J.; Estrella, B.; William, B.M.; Jeffrey, K.G.; Sempértegui, F. Assessment of the Binax NOW Streptococcus pneumoniae Urinary Antigen Test in Children with Nasopharyngeal Pneumococcal Carriaged. Clin. Infect. Dis. 2002, 34, 1025–1028. [Google Scholar] [CrossRef] [Green Version]
- Saukkoriipi, A.; Leskela, K.; Herva, E.; Leinonen, M. Streptococcus pneumoniae in nasopharyngeal secretions of healthy children: Comparison of real-time PCR and culture from STGG-transport medium. Mol. Cell Probes. 2004, 18, 147–153. [Google Scholar] [CrossRef]
- Pimenta, F.C.; Roundtree, A.; Soysal, A.; Bakir, M.; du Plessis, M.; Wolter, N.; von Gottberg, A.; McGee, L.; Carvalho Mda, G.; Beall, B. Sequential triplex real-time PCR assay for detecting 21 pneumococcal capsular serotypes that account for a high global disease burden. J. Clin. Microbiol. 2013, 51, 647–652. [Google Scholar] [CrossRef] [Green Version]
- Azzari, C.; Moriondo, M.; Indolfi, G.; Cortimiglia, M.; Canessa, C.; Becciolini, L.; Lippi, F.; de Martino, M.; Resti, M. Realtime PCR Is More Sensitive than Multiplex PCR for Diagnosis and Serotyping in Children with Culture Negative Pneumococcal Invasive Disease. PLoS ONE 2010, 5, e9282. [Google Scholar] [CrossRef] [Green Version]
- Kellner, J.D.; Scheifele, D.; Vanderkooi, O.G.; Macdonald, J.; Church, D.L.; Tyrrell, G.J. Effects of routine infant vaccination with the 7-valent pneumococcal conjugate vaccine on nasopharyngeal colonization with S. pneumoniae in children in Calgary, Canada. Pediatr. Infect. Dis. J. 2008, 27, 526–532. [Google Scholar] [CrossRef] [PubMed]
- Becker, K.; Skov, R.L.; von Eiff, C. Staphylococcus, Micrococcus, and Other Catalase-Positive Cocci. In Manuel of Clinical Microbiology, 11th ed.; Jorgensen, J.H., Carroll, K.C., Funke, G., Pfaller, M.A., Landry, M.L., Richter, S.S., Warnock, D.W., Eds.; American Society for Microbiology Press: Washington, DC, USA, 2015; Volume 1, pp. 354–382. [Google Scholar]
- Christensen, J.J.; Ruoff, K.L. Aerococcus, Abiotrophia, and Other Aerobic Catalase-Negative, Gram Positive Cocci. In Manual of Clinical Microbiology; Jorgensen, J.H., Carroll, K.C., Funke, G., Pfaller, M.A., Landry, M.L., Richter, S.S., Warnock, D.W., Eds.; American Society for Microbiology Press: Washington, DC, USA, 2015; Volume 1, pp. 422–436. [Google Scholar]
- Lingappa, J.R.; Lawrence, W.; West-Keefe, S.; Gautom, R.; Cookson, B.T. Diagnosis of community-acquired pertussis infection: Comparison of both culture and fluorescent-antibody assays with PCR detection using electrophoresis or dot blot hybridization. J. Clin. Microbiol. 2002, 40, 2908–2912. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Da Gloria Carvalho, M.; Tondella, M.L.; McCaustland, K.; Weidlich, L.; McGee, L.; Mayer, L.W.; Steigerwalt, A.; Whaley, M.; Facklam, R.R.; Fields, B.; et al. Evaluation and Improvement of Real-Time PCR Assays Targeting lytA, ply, and psaA Genes for Detection of Pneumococcal DNA. J. Clin. Microbiol. 2007, 45, 2460. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trzcinski, K.; Bogaert, D.; Wyllie, A.; Chu, M.L.; van der Ende, A.; Bruin, J.P.; van den Dobbelsteen, G.; Veenhoven, R.H.; Sanders, E.A. Superiority of trans-oral over trans-nasal sampling in detecting Streptococcus pneumoniae colonization in adults. PLoS ONE 2013, 8, e60520. [Google Scholar] [CrossRef] [Green Version]
- Tavares, D.A.; Handem, S.; Carvalho, R.J.; Paulo, A.C.; de Lencastre, H.; Hinds, J.; Sa-Leao, R. Identification of Streptococcus pneumoniae by a real-time PCR assay targeting SP2020. Sci. Rep. 2019, 9, 3285. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ganaie, F.; Maruhn, K.; Li, C.; Porambo, R.J.; Elverdal, P.L.; Abeygunwardana, C.; van der Linden, M.; Duus, J.; Sheppard, C.L.; Nahm, M.H. Structural, Genetic, and Serological Elucidation of Streptococcus pneumoniae Serogroup 24 Serotypes: Discovery of a New Serotype, 24C, with a Variable Capsule Structure. J. Clin. Microbiol. 2021, 59, e0054021. [Google Scholar] [CrossRef] [PubMed]
- Wyllie, A.L.; Rümke, L.W.; Arp, K.; Bosch, A.A.T.M.; Bruin, J.P.; Rots, N.Y.; Wijmenga-Monsuur, A.J.; Sanders, E.A.M.; Trzciński, K. Molecular surveillance on Streptococcus pneumoniae carriage in non-elderly adults; little evidence for pneumococcal circulation independent from the reservoir in children. Sci. Rep. 2016, 6, 34888. [Google Scholar] [CrossRef]
- Pasinato, A.; Indolfi, G.; Marchisio, P.; Valleriani, C.; Cortimiglia, M.; Spanevello, V.; Chiamenti, G.; Buzzetti, R.; Resti, M.; Azzari, C. Pneumococcal serotype distribution in 1315 nasopharyngeal swabs from a highly vaccinated cohort of Italian children as detected by RT-PCR. Vaccine 2014, 32, 1375–1381. [Google Scholar] [CrossRef]
- Ricketson, L.J.; Kellner, J.D. Invasive Pneumococcal Disease (IPD) Trends 1998 to Mid-2019 in Calgary, Canada: An Interrupted Time Series Analysis. In Proceedings of the International Syposium on Pneumococcus and Pneumococcal Disease, Toronto, ON, Canada, 19–23 June 2020; Abstract 104. Available online: https://cslide.ctimeetingtech.com/isppd20/attendee/confcal/session/calendar?q=Ricketson (accessed on 25 September 2020).
- Demczuk, W.; Griffith, A.; Montes, K.; Mallari, R.; Thorington, R.; Sawatzky, P.; Martin, I.; Mulvey, M. Laboratory Surveillance of Invasive Streptococcal Disease in Canada Annual Report 2018; Public Health Agency of Canada: Ottawa, ON, Canada, 2018; Available online: https://publications.gc.ca/collections/collection_2021/aspc-phac/HP57-4-2018-eng.pdf (accessed on 3 September 2021).
- Messaoudi, M.; Milenkov, M.; Albrich, W.C.; van der Linden, M.P.; Bénet, T.; Chou, M.; Sylla, M.; Barreto Costa, P.; Richard, N.; Klugman, K.P.; et al. The Relevance of a Novel Quantitative Assay to Detect up to 40 Major Streptococcus pneumoniae Serotypes Directly in Clinical Nasopharyngeal and Blood Specimens. PLoS ONE 2016, 11, e0151428. [Google Scholar] [CrossRef] [Green Version]
- Van Gils, E.J.; Veenhoven, R.H.; Hak, E.; Rodenburg, G.D.; Bogaert, D.; Ijzerman, E.P.; Bruin, J.P.; van Alphen, L.; Sanders, E.A. Effect of reduced-dose schedules with 7-valent pneumococcal conjugate vaccine on nasopharyngeal pneumococcal carriage in children: A randomized controlled trial. JAMA 2009, 302, 159–167. [Google Scholar] [CrossRef] [Green Version]
- Løvlie, A.; Vestrheim, D.F.; Aaberge, I.S.; Steens, A. Changes in pneumococcal carriage prevalence and factors associated with carriage in Norwegian children, four years after introduction of PCV13. BMC Infect. Dis. 2020, 20, 29. [Google Scholar] [CrossRef]
- Johnsborg, O.; Eldholm, V.; Bjørnstad, M.L.; Håvarstein, L.S. A predatory mechanism dramatically increases the efficiency of lateral gene transfer in Streptococcus pneumoniae and related commensal species. Mol. Microbiol. 2008, 69, 245–253. [Google Scholar] [CrossRef]
- Suzuki, N.; Yuyama, M.; Maeda, S.; Ogawa, H.; Mashiko, K.; Kiyoura, Y. Genotypic identification of presumptive S. pneumoniae by PCR using four genes highly specific for S. pneumoniae. J. Med. Microbiol. 2006, 55, 709–714. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hanage, W.P.; Kaijalainen, T.; Saukkoriipi, A.; Rickcord, J.L.; Spratt, B.G. A successful, diverse disease-associated lineage of nontypeable pneumococci that has lost the capsular biosynthesis locus. J. Clin. Microbiol. 2006, 44, 743–749. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shayegani, M.; Parsons, L.M.; Gibbons, W.E., Jr.; Campbell, D. Characterization of nontypable Streptococcus pneumoniae-like organisms isolated from outbreaks of conjunctivitis. J. Clin. Microbiol. 1982, 16, 8–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Richter, S.S.; Heilmann, K.P.; Dohrn, C.L.; Riahi, F.; Diekema, D.J.; Doern, G.V. Evaluation of pneumococcal serotyping by multiplex PCR and quellung reactions. J. Clin. Microbiol. 2013, 51, 4193–4195. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kellner, J.D.; Ricketson, L.J.; Demczuk, W.H.B.; Martin, I.; Tyrrell, G.J.; Vanderkooi, O.G.; Mulvey, M.R. Whole-Genome Analysis of Streptococcus pneumoniae Serotype 4 Causing Outbreak of Invasive Pneumococcal Disease, Alberta, Canada. Emerg. Infect. Dis. 2021, 27, 1867–1875. [Google Scholar] [CrossRef]
- Latifi-Navid, H.; Latifi-Navid, S.; Mostafaiy, B.; Jamalkandi, S.A.; Ahmadi, A. Pneumococcal Disease and the Effectiveness of the PPV23 Vaccine in Adults: A Two-Stage Bayesian Meta-Analysis of Observational and RCT Reports. Sci. Rep. 2018, 8, 11051. [Google Scholar] [CrossRef] [PubMed]
- Stacey, H.L.; Rosen, J.; Peterson, J.T.; Williams-Diaz, A.; Gakhar, V.; Sterling, T.M.; Acosta, C.J.; Nolan, K.M.; Li, J.; Pedley, A.; et al. Safety and immunogenicity of 15-valent pneumococcal conjugate vaccine (PCV-15) compared to PCV-13 in healthy older adults. Hum. Vaccines Immunother. 2019, 15, 530–539. [Google Scholar] [CrossRef] [Green Version]
- Hurley, D.; Griffin, C.; Young, M.; Scott, D.A.; Pride, M.W.; Scully, I.L.; Ginis, J.; Severs, J.; Jansen, K.U.; Gruber, W.C.; et al. Safety, Tolerability, and Immunogenicity of a 20-Valent Pneumococcal Conjugate Vaccine (PCV20) in Adults 60 to 64 Years of Age. Clin. Infect. Dis. 2021, 73, e1489–e1497. [Google Scholar] [CrossRef]
| lytA | piaA | SP2020 | Serotyping | Result |
|---|---|---|---|---|
| Pos a | Pos | N/D | Serotyped or Not typed | Positive |
| Pos | Neg | Pos | Serotyped or Not typed | Positive |
| Neg | Pos | Pos | Serotyped or Not typed | Positive |
| Pos | Neg | Neg | Serotyped | Positive |
| Neg | Pos | Neg | Serotyped | Positive |
| Pos | Neg | Neg | Not Typed | Equivocal |
| Neg | Pos | Neg | Not Typed | Equivocal |
| Neg | Neg | N/D | N/D | Negative |
| qPCR Results for All Samples n (%) | qPCR Results for Culture Positive Samples n (%) | qPCR Results for Culture Negative Samples n (%) | |
|---|---|---|---|
| PCV13/Related Serotype Carriage a | 61 (19.4) | 9 (12.7) | 52 (21.4) |
| NVT Carriage | 80 (25.5) | 21 (29.6) | 59 (24.3) |
| Multi-Carriage by NVT and PCV13-Serotypes | 31 (9.9) | 6 (8.4) | 25 (10.3) |
| Not Typed b | 142 (45.2) | 35 (49.3) | 107 (44.0) |
| Total Positive by qPCR | 314 (100.0) | 71 (100.0) | 243 (100.0) |
| qPCR Results for All Samples n (%) | qPCR Results for Culture Positive Samples n (%) | qPCR Results for Culture Negative Samples n (%) | |
|---|---|---|---|
| PCV13/Related Serotype Carriage a | 29 (17.1) | 11 (16.4) | 18 (17.5) |
| NVT Carriage | 60 (35.3) | 29 (43.3) | 31 (30.1) |
| Multi-Carriage by NVT and PCV13-Serotype | 8 (4.7) | 2 (3.0) | 6 (5.8) |
| Not Typed b | 73 (42.9) | 25 (37.3) | 48 (46.6) |
| Total Positive by qPCR | 170 (100.0) | 67 (100.0) | 103 (100.0) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ricketson, L.J.; Lidder, R.; Thorington, R.; Martin, I.; Vanderkooi, O.G.; Sadarangani, M.; Kellner, J.D. PCR and Culture Analysis of Streptococcus pneumoniae Nasopharyngeal Carriage in Healthy Children. Microorganisms 2021, 9, 2116. https://doi.org/10.3390/microorganisms9102116
Ricketson LJ, Lidder R, Thorington R, Martin I, Vanderkooi OG, Sadarangani M, Kellner JD. PCR and Culture Analysis of Streptococcus pneumoniae Nasopharyngeal Carriage in Healthy Children. Microorganisms. 2021; 9(10):2116. https://doi.org/10.3390/microorganisms9102116
Chicago/Turabian StyleRicketson, Leah J., Ravinder Lidder, Robyn Thorington, Irene Martin, Otto G. Vanderkooi, Manish Sadarangani, and James D. Kellner. 2021. "PCR and Culture Analysis of Streptococcus pneumoniae Nasopharyngeal Carriage in Healthy Children" Microorganisms 9, no. 10: 2116. https://doi.org/10.3390/microorganisms9102116
APA StyleRicketson, L. J., Lidder, R., Thorington, R., Martin, I., Vanderkooi, O. G., Sadarangani, M., & Kellner, J. D. (2021). PCR and Culture Analysis of Streptococcus pneumoniae Nasopharyngeal Carriage in Healthy Children. Microorganisms, 9(10), 2116. https://doi.org/10.3390/microorganisms9102116






