Fungi Dominated the Incorporation of 13C-CO2 into Microbial Biomass in Tomato Rhizosphere Soil under Different CO2 Concentrations
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Site
2.2. Experimental Set-Up
2.3. Analysis of Biochemical Properties
2.4. PLFA Analysis
2.5. Statistical Analysis
3. Results
3.1. Effect of CO2 Concentration on the Tomato Biomass and AT% 13C/12C
3.2. Effect of CO2 Concentration on the Biochemical Properties of the Soils
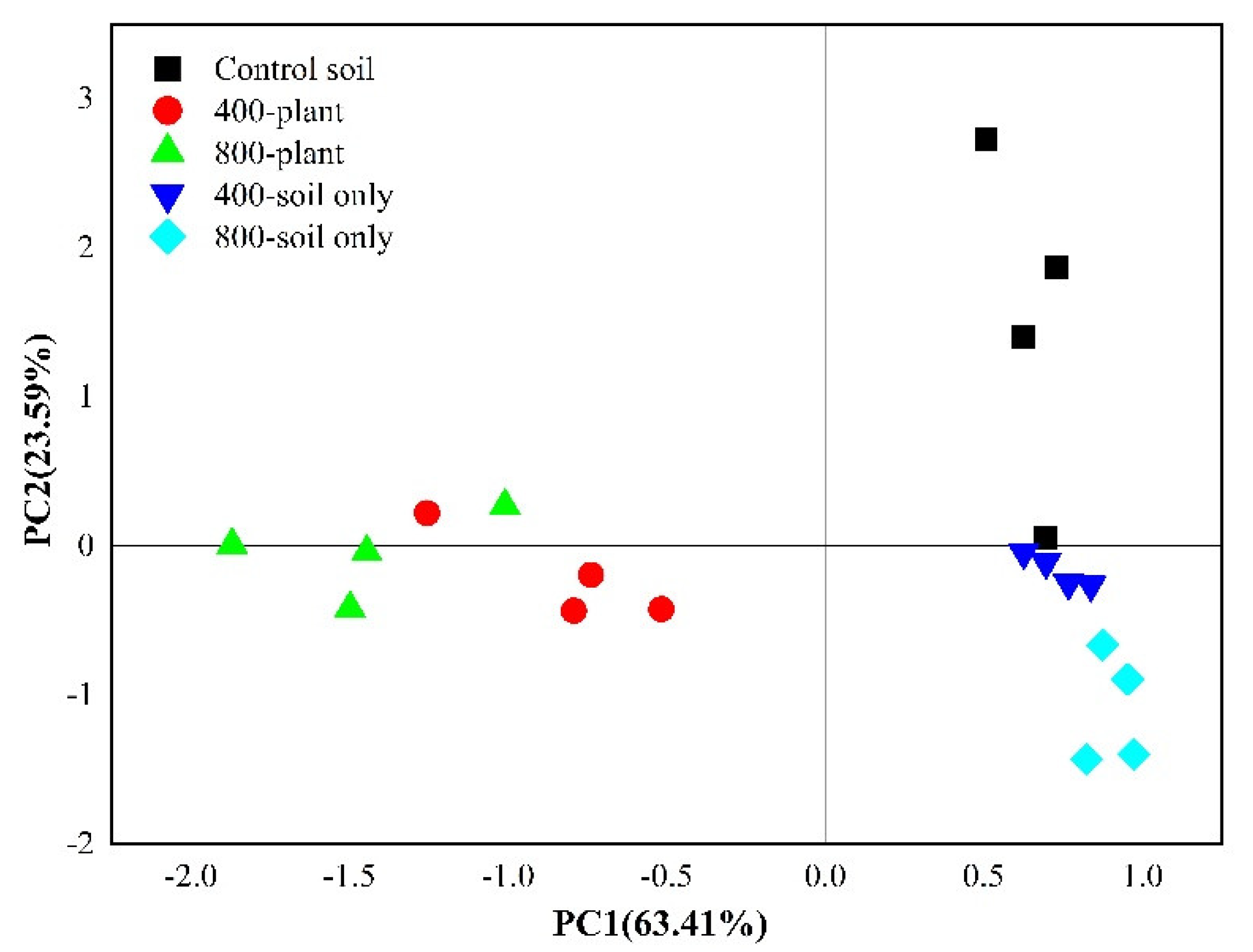
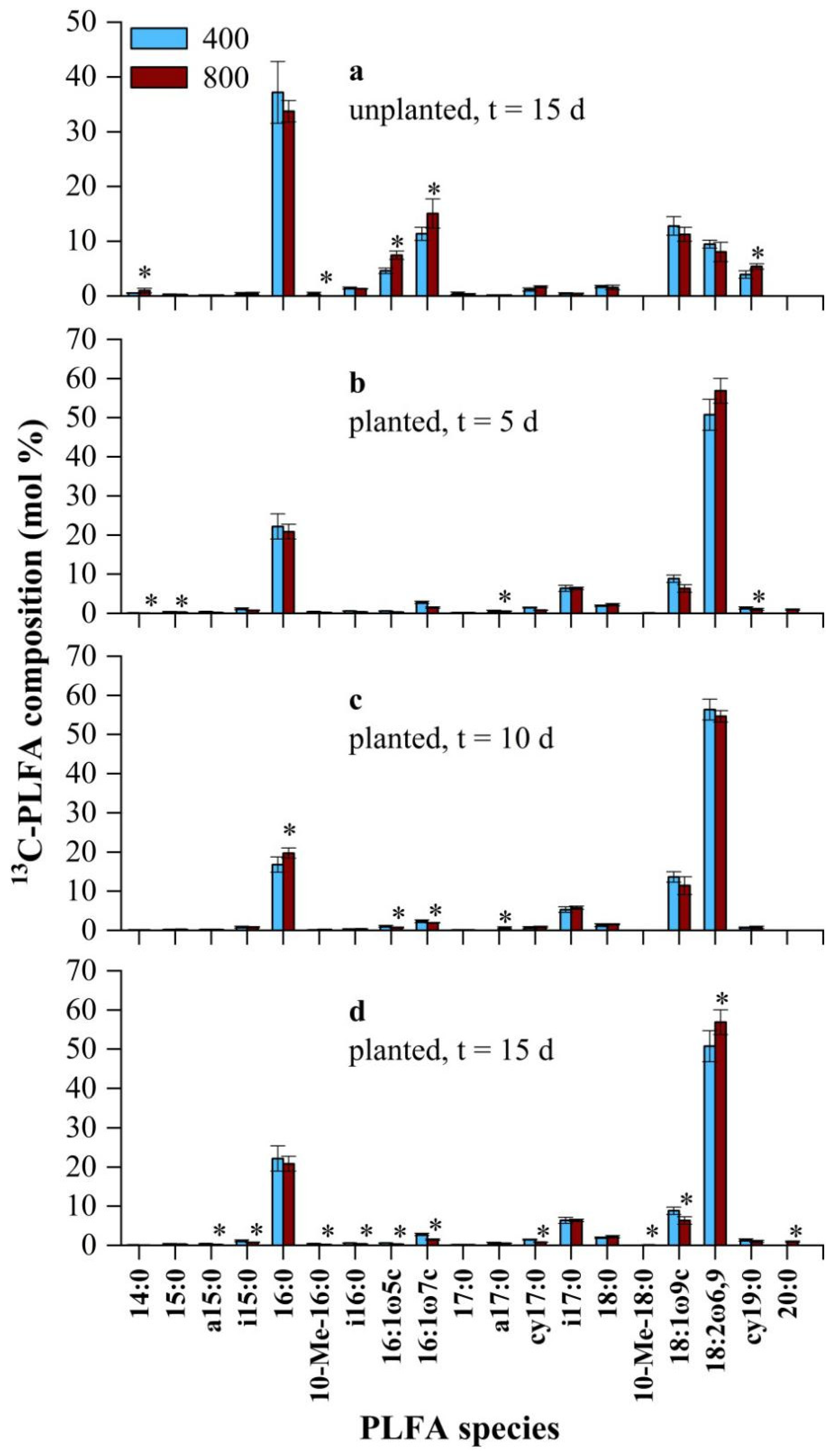
3.3. Effect of CO2 Concentration on the Soil Microbial Community
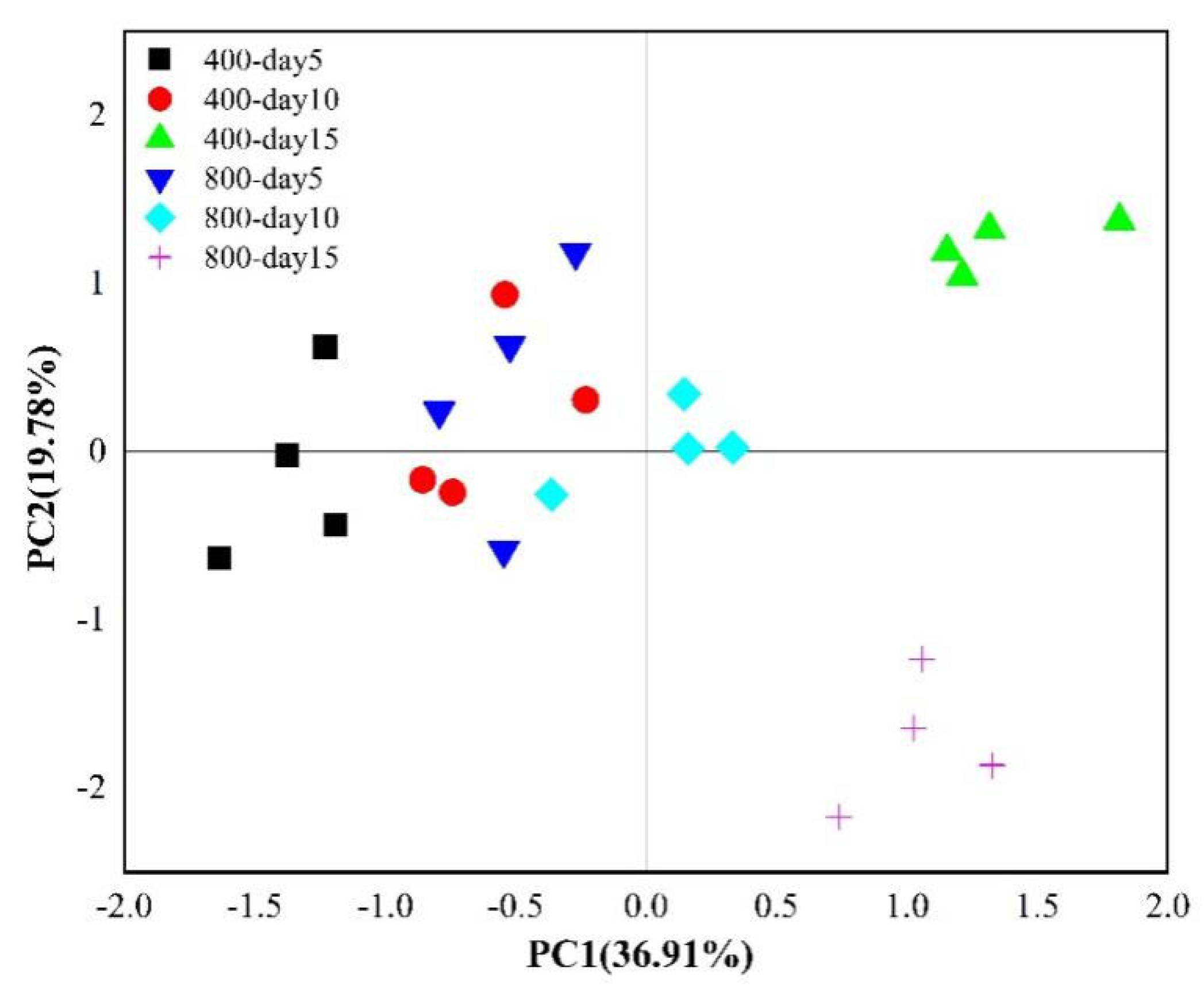
3.4. Variation in Soil Microbial Composition Incorporating 13C-Rhizodeposition under Different CO2 Concentrations
3.5. Relationship between PLFAs and Environmental Factors
4. Discussion
4.1. Effect of CO2 Concentration on 13C-Plant Biomass
4.2. Effect of CO2 Concentration on Microbial Community Structure
4.3. Effect of CO2 Concentration on 13C-Microbial Community Structure
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- IPCC. IPCC, 2013: Climate Change 2013: The Physical Science Basis. Contribution of Working Group I to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change; Cambridge University Press: Cambridge, UK; New York, NY, USA, 2014; 1535p. Available online: trove.nla.gov.au/work/190076309 (accessed on 24 August 2021).
- Procter, A.C.; Gill, R.A.; Fay, P.A.; Polley, H.W.; Jackson, R.B. Soil carbon responses to past and future CO2 in three Texas prairie soils. Soil Biol. Biochem. 2015, 83, 66–75. [Google Scholar] [CrossRef] [Green Version]
- Van der Kooi, C.J.; Reich, M.; Löw, M.; De Kok, L.J.; Tausz, M. Growth and yield stimulation under elevated CO2 and drought: A meta-analysis on crops. Environ. Exp. Bot. 2016, 122, 150–157. [Google Scholar] [CrossRef]
- Calvo, O.C.; Franzaring, J.; Schmid, I.; Fangmeier, A. Root exudation of carbohydrates and cations from barley in response to drought and elevated CO2. Plant Soil 2019, 438, 127–142. [Google Scholar] [CrossRef]
- Saha, S.; Sehgal, V.K.; Chakraborty, D.; Pal, M. Atmospheric carbon dioxide enrichment induced modifications in canopy radiation utilization, growth and yield of chickpea (Cicer arietinum L.). Agric. For. Meteorol. 2015, 202, 102–111. [Google Scholar] [CrossRef]
- Nguyen, C. Rhizodeposition of organic C by plants: Mechanisms and controls. In Sustainable Agriculture; Springer: Dordrecht, The Netherlands, 2003; pp. 97–123. [Google Scholar] [CrossRef]
- Bertin, C.; Yang, X.; Weston, L.A. The role of root exudates and allelochemicals in the rhizosphere. Plant Soil 2003, 256, 67–83. [Google Scholar] [CrossRef]
- Li, X.; Dong, J.; Chu, W.; Chen, Y.; Duan, Z. The relationship between root exudation properties and root morphological traits of cucumber grown under different nitrogen supplies and atmospheric CO2 concentrations. Plant Soil 2018, 425, 415–432. [Google Scholar] [CrossRef]
- Badri, D.V.; Vivanco, J.M. Regulation and function of root exudates. Plant Cell Environ. 2010, 32, 666–681. [Google Scholar] [CrossRef]
- Wu, Q.; Congzhi, Z.; Zhenghong, Y.; Jiabao, Z.; Chunwu, Z.; Zhanhui, Z.; Jiananran, X.; Jinlin, C. Effects of elevated CO2 and nitrogen addition on organic carbon and aggregates in soil planted with different rice cultivars. Plant Soil 2018, 432, 245–258. [Google Scholar] [CrossRef]
- Yu, Z.; Li, Y.; Wang, G.; Liu, J.; Liu, J.; Liu, X.; Herbert, S.J.; Jin, J. Effectiveness of elevated CO2 mediating bacterial communities in the soybean rhizosphere depends on genotypes. Agric. Ecosyst. Environ. 2016, 231, 229–232. [Google Scholar] [CrossRef]
- Runion, G.B.; Curl, E.A.; Rogers, H.H.; Backman, P.A.; Rodríguez-Kábana, R.; Helms, B.E. Effects of free-air CO2 enrichment on microbial populations in the rhizosphere and phyllosphere of cotton. Agric. For. Meteorol. 1994, 70, 117–130. [Google Scholar] [CrossRef]
- Wang, P.; Marsh, E.L.; Ainsworth, E.A.; Leakey, A.D.B.; Sheflin, A.M.; Schachtman, D.P. Shifts in microbial communities in soil, rhizosphere and roots of two major crop systems under elevated CO2 and O3. Sci. Rep. 2017, 7, 15019. [Google Scholar] [CrossRef] [Green Version]
- Blagodatskaya, E.; Blagodatsky, S.; Dorodnikov, M.; Kuzyakov, Y. Elevated atmospheric CO2 increases microbial growth rates in soil: Results of three CO2 enrichment experiments. Glob. Chang. Biol. 2010, 16, 836–848. [Google Scholar] [CrossRef] [Green Version]
- Qiu, Y.; Jiang, Y.; Guo, L.; Zhang, L.; Hu, S. Shifts in the Composition and Activities of Denitrifiers Dominate CO2 Stimulation of N2O Emissions. Environ. Sci. Technol. 2019, 53, 11204–11213. [Google Scholar] [CrossRef]
- Treonis, A.M.; Ostle, N.J.; Stott, A.W.; Primrose, R.; Grayston, S.J.; Ineson, P. Identification of groups of metabolically-active rhizosphere microorganisms by stable isotope probing of PLFAs. Soil Biol. Biochem. 2004, 36, 533–537. [Google Scholar] [CrossRef]
- Esperschütz, J.; Gattinger, A.; Buegger, F.; Lang, H.; Munch, J.C.; Winkler, M.S.B. A continuous labelling approach to recover photosynthetically fixed carbon in plant tissue and rhizosphere organisms of young beech trees (Fagus sylvatica L.) using 13C depleted CO2. Plant Soil 2009, 323, 21–29. [Google Scholar] [CrossRef]
- Zhang, Q.; Jin, H.; Zhou, H.; Cai, M.; Li, Y.; Zhang, G.; Di, H. Variation of soil anaerobic microorganisms connected with anammox processes by 13C-phospholipid fatty acid analysis among long-term fertilization regimes in a crop rotation system. Appl. Soil Ecol. 2018, 133, 34–43. [Google Scholar] [CrossRef]
- Da Costa, D.P.; Dias, A.C.F.; Cotta, S.R.; Vilela, D.; de Andrade, P.A.M.; Pellizari, V.H.; Andreote, F.D. Changes of bacterial communities in the rhizosphere of sugarcane under elevated concentration of atmospheric CO2. GCB Bioenergy 2018, 10, 137–145. [Google Scholar] [CrossRef] [Green Version]
- Mayr, C.; Miller, M.; Insam, H. Elevated CO2 alters community-level physiological profiles and enzyme activities in alpine grassland. J. Microbiol. Methods 1999, 36, 35–43. [Google Scholar] [CrossRef]
- Grayston, S.J.; Campbell, C.D.; Lutze, J.L.; Gifford, R.M. Impact of elevated CO2 on the metabolic diversity of microbial communities in N-limited grass swards. Plant Soil 1998, 203, 289–300. [Google Scholar] [CrossRef]
- Diao, T.; Peng, Z.; Niu, X.; Yang, R.; Ma, F.; Guo, L. Changes of Soil Microbes Related with Carbon and Nitrogen Cycling after Long-Term CO2 Enrichment in a Typical Chinese Maize Field. Sustainability 2020, 12, 1250. [Google Scholar] [CrossRef] [Green Version]
- Fang, H.; Cheng, S.; Lin, E.; Yu, G.; Niu, S.; Wang, Y.; Xu, M.; Dang, X.; Li, L.; Wang, L. Elevated atmospheric carbon dioxide concentration stimulates soil microbial activity and impacts water-extractable organic carbon in an agricultural soil. Biogeochemistry 2015, 122, 253–267. [Google Scholar] [CrossRef]
- Zheng, J.-Q.; Han, S.-J.; Zhou, Y.-M.; Ren, F.-R.; Xin, L.-H.; Zhang, Y. Microbial Activity in a Temperate Forest Soil as Affected by Elevated Atmospheric CO2. Pedosphere 2010, 20, 427–435. [Google Scholar] [CrossRef]
- Cheng, L.; Booker, F.L.; Burkey, K.O.; Tu, C.; Shew, H.D.; Rufty, T.W.; Fiscus, E.L.; Deforest, J.L.; Hu, S. Soil microbial responses to elevated CO2 and O3 in a nitrogen-aggrading agroecosystem. PLoS ONE 2011, 6, e21377. [Google Scholar] [CrossRef] [Green Version]
- Haase, S.; Philippot, L.; Neumann, G.; Marhan, S.; Kandeler, E. Local response of bacterial densities and enzyme activities to elevated atmospheric CO2 and different N supply in the rhizosphere of Phaseolus vulgaris L. Soil Biol. Biochem. 2008, 40, 1225–1234. [Google Scholar] [CrossRef]
- Wang, H.; Fan, H.; Yao, H. Effects of Elevated CO2 on Tomato (Lycopersicon esculentum Mill.) Growth and Rhizosphere Soil Microbial Community Structure and Functionality. Agronomy 2020, 10, 1752. [Google Scholar] [CrossRef]
- Wu, W.X.; Liu, W.; Lu, H.H.; Chen, Y.X.; Devare, M.; Thies, J. Use of 13C labeling to assess carbon partitioning in transgenic and nontransgenic (parental) rice and their rhizosphere soil microbial communities. FEMS Microbiol. Ecol. 2009, 67, 93–102. [Google Scholar] [CrossRef]
- Schonhof, I.; Kläring, H.P.; Krumbein, A.; Schreiner, M. Interaction Between Atmospheric CO2 and Glucosinolates in Broccoli. J. Chem. Ecol. 2007, 33, 105–114. [Google Scholar] [CrossRef]
- Long, S.P.; Ainsworth, E.; Rogers, A.; Ort, D.R. Rising atmospheric carbon dioxide: Plants FACE the future. Annu. Rev. Plant Biol. 2004, 55, 591–628. [Google Scholar] [CrossRef]
- Korres, N.E.; Norsworthy, J.K.; Tehranchian, P.; Gitsopoulos, T.K.; Loka, D.A.; Oosterhuis, D.M.; Gealy, D.R.; Moss, S.R.; Burgos, N.R.; Miller, M.R. Cultivars to face climate change effects on crops and weeds: A review. Agron. Sustain. Dev. 2016, 36, 12. [Google Scholar] [CrossRef] [Green Version]
- Butterly, C.R.; Wang, X.; Armstrong, R.D.; Chen, D.; Tang, C. Elevated CO2 induced rhizosphere effects on the decomposition and N recovery from crop residues. Plant Soil 2016, 408, 55–71. [Google Scholar] [CrossRef]
- Xu, Q.; O’Sullivan, J.B.; Wang, X.; Tang, C. Elevated CO2 alters the rhizosphere effect on crop residue decomposition. Plant Soil 2019, 436, 413–426. [Google Scholar] [CrossRef]
- Ferrocino, I.; Chitarra, W.; Pugliese, M.; Gilardi, G.; Gullino, M.L.; Garibaldi, A. Effect of elevated atmospheric CO2 and temperature on disease severity of Fusarium oxysporum f.sp. lactucae on lettuce plants. Appl. Soil Ecol. 2013, 72, 1–6. [Google Scholar] [CrossRef]
- Martinsen, V.; Austrheim, G.; Mysterud, A.; Mulder, J. Effects of herbivory on N-cycling and distribution of added 15NH4+ in N-limited low-alpine grasslands. Plant Soil 2011, 347, 279–292. [Google Scholar] [CrossRef]
- Yao, H.; Thornton, B.; Paterson, E. Incorporation of 13C-labelled rice rhizodeposition carbon into soil microbial communities under different water status. Soil Biol. Biochem. 2012, 53, 72–77. [Google Scholar] [CrossRef]
- Bligh, E.G.; Dyer, W.J. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 1959, 37, 911–917. [Google Scholar] [CrossRef] [Green Version]
- Thornton, B.; Zhang, Z.; Mayes, R.W.; Hogberg, M.N.; Midwood, A.J. Can gas chromatography combustion isotope ratio mass spectrometry be used to quantify organic compound abundance? Rapid Commun. Mass Spectrom. 2011, 25, 2433–2438. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Ding, W.; Yu, H.; He, X. Carbon uptake by a microbial community during 30-day treatment with 13C-glucose of a sandy loam soil fertilized for 20 years with NPK or compost as determined by a GC–C–IRMS analysis of phospholipid fatty acids. Soil Biol. Biochem. 2013, 57, 228–236. [Google Scholar] [CrossRef]
- Frostegård, Å.; Tunlid, A.; Bååthb, E. Use and misuse of PLFA measurements in soils. Soil Biol. Biochem. 2011, 43, 1621–1625. [Google Scholar] [CrossRef]
- Lai, J. Quantitative Ecology: Applications of the R Language; Higher Education Press: Beijing, China, 2014. [Google Scholar]
- Canarini, A.; Wanek, W.; Merchant, A.; Richter, A.; Kaiser, C. Root exudation of primary metabolites: Mechanisms and their roles in plant responses to environmental stimuli. Front. Plant Sci. 2019, 10, 157. [Google Scholar] [CrossRef] [Green Version]
- Hodge, A.; Paterson, E.; Grayston, S.J.; Campbell, C.; Ord, B.G.; Killham, K. Characterization and microbial utilization of exudate material from the rhizosphere of Lolium perenne grown under CO2 enrichment. Soil Biol. Biochem. 1998, 583, 1033–1043. [Google Scholar] [CrossRef]
- Zak, D.R.; Pregitzer, K.S.; Curtis, P.S.; Holmes, W.E. Atmospheric CO2 and the composition and function of soil microbial communities. Ecol. Appl. 2000, 10, 47–59. [Google Scholar] [CrossRef] [Green Version]
- Guenet, B.; Lenhart, K.; Leloup, J.; Giusti-Miller, S.; Pouteau, V.; Mora, P.; Nunan, N.; Abbadie, L. The impact of long-term CO2 enrichment and moisture levels on soil microbial community structure and enzyme activities. Geoderma 2012, 170, 331–336. [Google Scholar] [CrossRef]
- Hayden, H.L.; Mele, P.M.; Bougoure, D.S.; Allan, C.Y.; Norng, S.; Piceno, Y.M.; Brodie, E.L.; DeSantis, T.Z.; Andersen, G.L.; Williams, A.L. Changes in the microbial community structure of bacteria, archaea and fungi in response to elevated CO2 and warming in an Australian native grassland soil. Environ. Microbiol. 2012, 14, 3081–3096. [Google Scholar] [CrossRef]
- Long, X.E.; Yao, H.; Huang, Y.; Wei, W.; Zhu, Y.G. Phosphate levels influence the utilization of rice rhizodeposition carbon and the phosphate-solubilising microbial community in a paddy soil. Soil Biol. Biochem. 2018, 118, 103–114. [Google Scholar] [CrossRef]
- Klamer, M.; Bååthb, E. Estimation of conversion factors for fungal biomass determination in compost using ergosterol and PLFA 18:2ω6,9. Soil Biol. Biochem. 2004, 36, 57–65. [Google Scholar] [CrossRef]
- Cheng, L.; Booker, F.L.; Tu, C.; Burkey, K.O.; Zhou, L.; Shew, H.D.; Rufty, T.W.; Hu, S. Arbuscular mycorrhizal fungi increase organic carbon decomposition under elevated CO2. Science 2012, 337, 1084–1087. [Google Scholar] [CrossRef]
- Lesaulnier, C.; Papamichail, D.; McCorkle, S.; Ollivier, B.; Skiena, S.; Taghavi, S.; Zak, D.; Van Der Lelie, D. Elevated atmospheric CO2 affects soil microbial diversity associated with trembling aspen. Environ. Microbiol. 2008, 10, 926–941. [Google Scholar] [CrossRef] [Green Version]
- Gill, R.A.; Polley, H.W.; Johnson, H.B.; Anderson, L.J.; Maherali, H.; Jackson, R.B. Nonlinear grassland responses to past and future atmospheric CO2. Nature 2002, 417, 279–282. [Google Scholar] [CrossRef]
- Hu, S.; Chapin, F.S.; Firestone, M.; Field, C.; Chiariello, N. Nitrogen limitation of microbial decomposition in a grassland under elevated CO2. Nature 2001, 409, 188–191. [Google Scholar] [CrossRef]
- Ho, A.; Di Lonardo, D.P.; Bodelier, P.L.E. Revisiting life strategy concepts in environmental microbial ecology. FEMS Microbiol. Ecol. 2017, 93, fix006. [Google Scholar] [CrossRef] [Green Version]
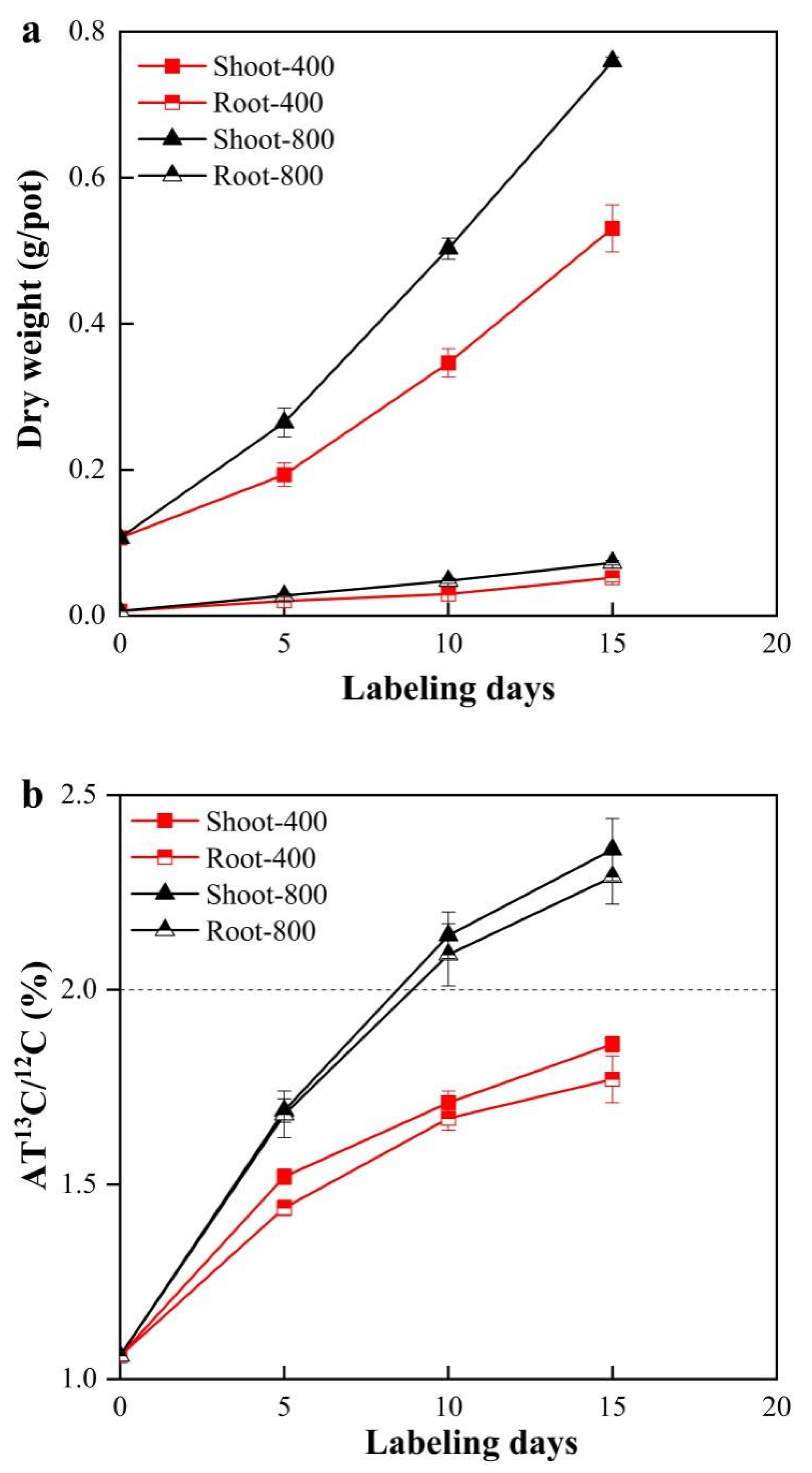

| Labeled Days | CO2 Concentration (µmol·mol−1) | Labeled Amount (nmol·g −1) Average ± STD | Enrichment Rate (%) Average ± STD |
|---|---|---|---|
| 5 | 400 1 | 24.43 ± 4.39 ab 2 | 7.43 ± 1.35 b |
| 800 | 60.05 ± 11.37 b | 15.13 ± 1.45 c | |
| 10 | 400 | 49.66 ± 9.6 ab | 12.65 ± 1.95 c |
| 800 | 150.46 ± 45.25 c | 34.27 ± 6.29 d | |
| 15 | 400 | 188.71 ± 16.14 c | 35.82 ± 2.33 d |
| 800 | 456.76 ± 67.91 d | 68.89 ± 4.25 e | |
| CO2 concentrations | * | ||
| Labeling days | *** | ||
| CO2 concentration:Labeling days | ns | ||
| Unplanted soil | 400 | 3.52 ± 0.45 a | 1.08 ± 0.14 a |
| 800 | 5.61 ± 0.79 a | 1.65 ± 0.22 a |
| Factors | PLFA Composition | 13C-PLFA Composition | ||
|---|---|---|---|---|
| Mantel Statistic r | Significance | Mantel Statistic r | Significance | |
| CO2 | 0.3887 | 0.0562 | 0.4556 | 0.0289 * |
| DOC 1 | 0.3636 | 0.0489 * | 0.4281 | 0.0256 * |
| TDW | 0.4905 | 0.0204 * | 0.7088 | 0.0008 *** |
| CO2_DOC | 0.1648 | 0.1881 | 0.1958 | 0.1594 |
| CO2_TDW | −0.0714 | 0.6301 | −0.4199 | 0.9494 |
| DOC_TDW | −0.02641 | 0.5555 | −0.2638 | 0.846 |
| TDW_DOC | 0.3543 | 0.0685 | 0.6581 | 0.0035 ** |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, H.; Wang, J.; Ge, C.; Yao, H. Fungi Dominated the Incorporation of 13C-CO2 into Microbial Biomass in Tomato Rhizosphere Soil under Different CO2 Concentrations. Microorganisms 2021, 9, 2121. https://doi.org/10.3390/microorganisms9102121
Wang H, Wang J, Ge C, Yao H. Fungi Dominated the Incorporation of 13C-CO2 into Microbial Biomass in Tomato Rhizosphere Soil under Different CO2 Concentrations. Microorganisms. 2021; 9(10):2121. https://doi.org/10.3390/microorganisms9102121
Chicago/Turabian StyleWang, Hehua, Juan Wang, Chaorong Ge, and Huaiying Yao. 2021. "Fungi Dominated the Incorporation of 13C-CO2 into Microbial Biomass in Tomato Rhizosphere Soil under Different CO2 Concentrations" Microorganisms 9, no. 10: 2121. https://doi.org/10.3390/microorganisms9102121
APA StyleWang, H., Wang, J., Ge, C., & Yao, H. (2021). Fungi Dominated the Incorporation of 13C-CO2 into Microbial Biomass in Tomato Rhizosphere Soil under Different CO2 Concentrations. Microorganisms, 9(10), 2121. https://doi.org/10.3390/microorganisms9102121






