Identification of Potential Citrate Metabolism Pathways in Carnobacterium maltaromaticum
Abstract
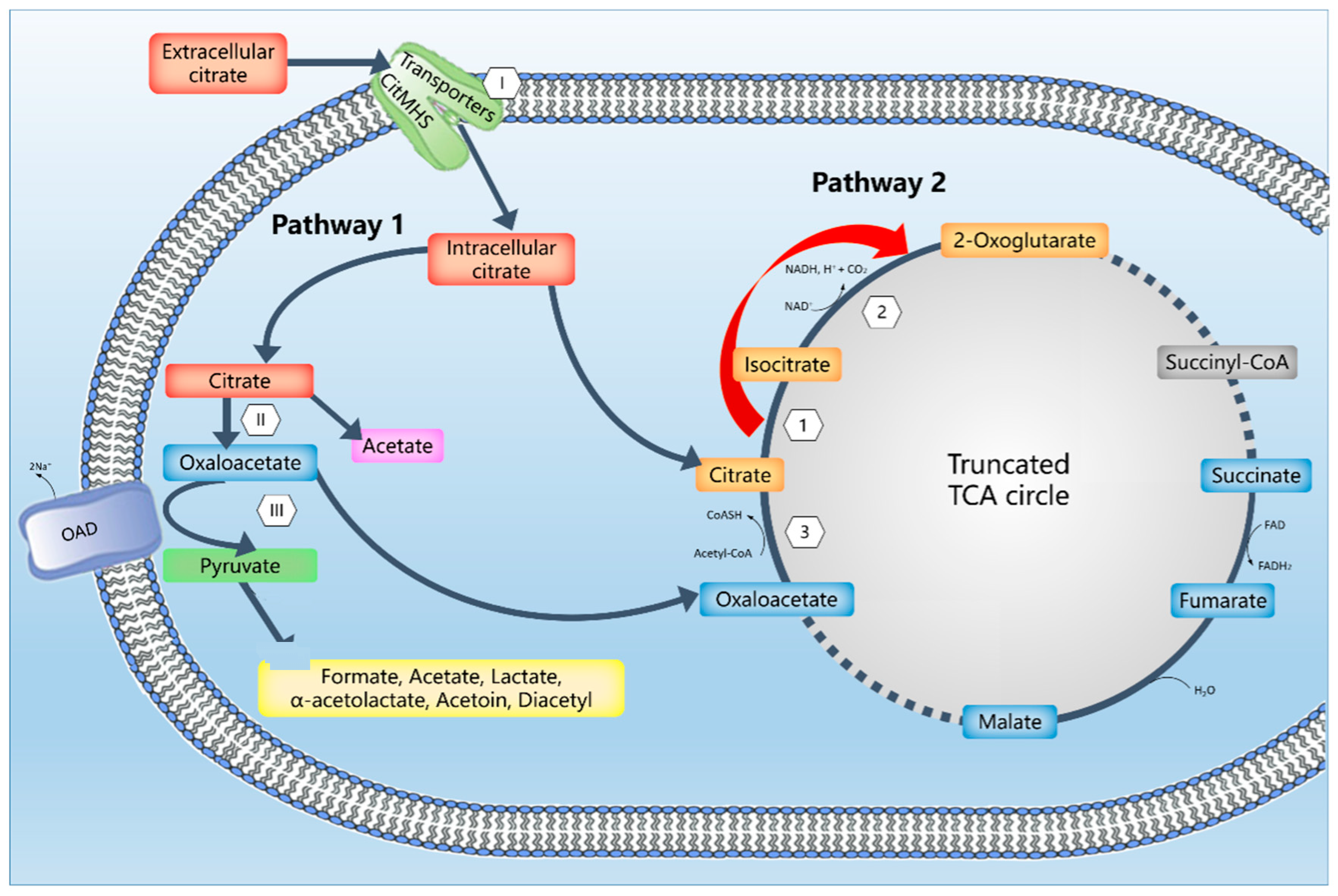
:1. Introduction
2. Materials and Methods
2.1. Strains Collection, Phenotypic Tests, and Bioinformatic Analyses
2.2. Phenotypic Tests for Citrate Metabolism and Test for Growth on a Citrate-Containing Medium
2.3. Whole-Genome Sequencing
2.4. Bioinformatic Analyses for Genome Annotation
3. Results and Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hugenholtz, J. Citrate metabolism in lactic acid bacteria. FEMS Microbiol. Rev. 1993, 12, 165–178. [Google Scholar] [CrossRef]
- Morea, M.; Baruzzi, F.; Cocconcelli, P. Molecular and physiological characterization of dominant bacterial populations in traditional Mozzarella cheese processing. J. Appl. Microbiol. 1999, 87, 574–582. [Google Scholar] [CrossRef]
- Palles, T.; Beresford, T.; Condon, S.; Cogan, T.M. Citrate metabolism in Lactobacillus casei and Lactobacillus plantarum. J. Appl. Microbiol. 1998, 85, 147–154. [Google Scholar] [CrossRef]
- Bintsis, T. Lactic acid Bacteria as starter cultures: An update in their metabolism and genetics. AIMS Microbiol. 2018, 4, 665–684. [Google Scholar] [CrossRef] [PubMed]
- Leisner, J.J.; Laursen, B.G.; Prévost, H.; Drider, D.; Dalgaard, P. Carnobacterium: Positive and negative effects in the environment and in foods. FEMS Microbiol. Rev. 2007, 31, 592–613. [Google Scholar] [CrossRef] [Green Version]
- Leisner, J.J.; Hansen, M.A.; Larsen, M.H.; Hansen, L.; Ingmer, H.; Sørensen, S.J. The genome sequence of the lactic acid bacterium, Carnobacterium maltaromaticum ATCC 35586 encodes potential virulence factors. Int. J. Food Microbiol. 2012, 152, 107–115. [Google Scholar] [CrossRef]
- Toranzo, A.; Romalde, J.; Nunez, S.; Figueras Huerta, A.; Barja, J. An epizootic in farmed, market-sized rainbow trout in Spain caused by a strain of Carnobactenum piscicola of unusual virulence. Dis. Aquat. Org. 1993, 121, 6107–6108. [Google Scholar] [CrossRef]
- Roh, H.; Kim, B.S.; Lee, M.K.; Park, C.-I.; Kim, D.-H. Genome-wide comparison of Carnobacterium maltaromaticum derived from diseased fish harbouring important virulence-related genes. J. Fish Dis. 2020, 43, 1029–1037. [Google Scholar] [CrossRef]
- Afzal, M.I.; Ariceaga, C.C.G.; Lhomme, E.; Ali, N.K.; Payot, S.; Burgain, J.; Gaiani, C.; Borges, F.; Revol-Junelles, A.-M.; Delaunay, S.; et al. Characterization of Carnobacterium maltaromaticum LMA 28 for its positive technological role in soft cheese making. Food Microbiol. 2013, 36, 223–230. [Google Scholar] [CrossRef] [PubMed]
- Østlie, H.M.; Treimo, J.; Narvhus, J.A. Effect of temperature on growth and metabolism of probiotic bacteria in milk. Int. Dairy J. 2005, 15, 989–997. [Google Scholar] [CrossRef]
- Singh, T.; Drake, M.A.; Cadwallader, K.R. Flavor of Cheddar cheese: A chemical and sensory perspective. Compr. Rev. Food Sci. Food Saf. 2003, 2, 166–189. [Google Scholar] [CrossRef]
- García-Quintáns, N.; Blancato, V.S.; Repizo, G.D.; Magni, C.; López, P. Citrate metabolism and aroma compound production in lactic acid bacteria. In Molecular Aspects of Lactic Acid Bacteria for Traditional and New Applications; Research Signpost: Trivandrum, India, 2008. [Google Scholar]
- Leisner, J.J.; Vogensen, F.K.; Kollmann, J.; Aideh, B.; Vandamme, P.; Vancanneyt, M.; Ingmer, H. α-Chitinase activity among lactic acid bacteria. Syst. Appl. Microbiol. 2008, 31, 151–156. [Google Scholar] [CrossRef]
- Kempler, G.M.; McKay, L.L. Improved medium for detection of citrate-fermenting Streptococcus lactis subsp. diacetylactis. Appl. Environ. Microbiol. 1980, 39, 926–927. [Google Scholar] [CrossRef] [Green Version]
- Smart, K.F.; Aggio, R.B.M.; Van Houtte, J.R.; Villas-Bôas, S.G. Analytical platform for metabolome analysis of microbial cells using methyl chloroformate derivatization followed by gas chromatography-mass spectrometry. Nat. Protoc. 2010, 5, 1709–1729. [Google Scholar] [CrossRef]
- Johnsen, L.G.; Skou, P.B.; Khakimov, B.; Bro, R. Gas chromatography–mass spectrometry data processing made easy. J. Chromatogr. A 2017, 1503, 57–64. [Google Scholar] [CrossRef] [PubMed]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M. The RAST Server: Rapid annotations using subsystems technology. BMC Genom. 2008, 9, 75–content. [Google Scholar] [CrossRef] [Green Version]
- Overbeek, R.; Olson, R.; Pusch, G.D.; Olsen, G.J.; Davis, J.J.; Disz, T.; Edwards, R.A.; Gerdes, S.; Parrello, B.; Shukla, M.; et al. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Res. 2014, 42, 206–214. [Google Scholar] [CrossRef] [PubMed]
- Brettin, T.; Davis, J.J.; Disz, T.; Edwards, R.A.; Gerdes, S.; Olsen, G.J.; Olson, R.; Overbeek, R.; Parrello, B.; Pusch, G.D.; et al. RASTtk: A modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci. Rep. 2015, 5, 8365. [Google Scholar] [CrossRef] [Green Version]
- Letunic, I.; Bork, P. Interactive tree of life (iTOL) v3: An online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016, 44, 242–245. [Google Scholar] [CrossRef] [PubMed]
- Schultz, J.; Copley, R.R.; Doerks, T.; Ponting, C.P.; Bork, P. SMART: A web-based tool for the study of genetically mobile domains. Nucleic Acids Res. 2000, 28, 231–234. [Google Scholar] [CrossRef] [PubMed]
- El-Gebali, S.; Mistry, J.; Bateman, A.; Eddy, S.R.; Luciani, A.; Potter, S.C.; Qureshi, M.; Richardson, L.J.; Salazar, G.A.; Smart, A. The Pfam protein families database in 2019. Nucleic Acids Res. 2019, 47, 427–432. [Google Scholar] [CrossRef]
- Consortium, U. UniProt: A hub for protein information. Nucleic Acids Res. 2015, 43, 204–212. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M. The KEGG Database Novartis Foundation Symposium. 2002, 247, 91–100.
- Vallenet, D.; Belda, E.; Calteau, A.; Cruveiller, S.; Engelen, S.; Lajus, A.; Le Fèvre, F.; Longin, C.; Mornico, D.; Roche, D.; et al. MicroScope—An integrated microbial resource for the curation and comparative analysis of genomic and metabolic data. Nucleic Acids Res. 2013, 41, 636–647. [Google Scholar] [CrossRef]
- Petkau, A.; Stuart-Edwards, M.; Stothard, P.; Van Domselaar, G. Interactive microbial genome visualization with GView. Bioinformatics 2010, 26, 3125–3126. [Google Scholar] [CrossRef] [PubMed]
- Krogh, A.; Larsson, B.; Von Heijne, G.; Sonnhammer, E.L. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 2001, 305, 567–580. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iskandar, C.F.; Borges, F.; Taminiau, B.; Daube, G.; Zagorec, M.; Remenant, B.; Leisner, J.J.; Hansen, M.A.; Sørensen, S.J.; Mangavel, C.; et al. Comparative genomic analysis reveals ecological differentiation in the genus Carnobacterium. Front. Microbiol. 2017, 8, 357. [Google Scholar] [CrossRef] [Green Version]
- Eisenbach, L.; Geissler, A.J.; Ehrmann, M.A.; Vogel, R.F. Comparative genomics of Lactobacillus sakei supports the development of starter strain combinations. Microbiol. Res. 2019, 221, 1–9. [Google Scholar] [CrossRef]
- Blancato, V.S.; Pagliai, F.A.; Magni, C.; Gonzalez, C.F.; Lorca, G.L. Functional analysis of the citrate activator CitO from Enterococcus faecalis implicates a divalent metal in ligand binding. Front. Microbiol. 2016, 7, 101. [Google Scholar] [CrossRef]
- Blancato, V.S.; Repizo, G.D.; Suárez, C.A.; Magni, C. Transcriptional regulation of the citrate gene cluster of Enterococcus faecalis Involves the GntR family transcriptional activator CitO. J. Bacteriol. 2008, 190, 7419–7430. [Google Scholar] [CrossRef] [Green Version]
- Lensbouer, J.J.; Doyle, R.P. Secondary transport of metal–citrate complexes: The CitMHS family. Crit. Rev. Biochem. Mol. Biol. 2010, 45, 453–462. [Google Scholar] [CrossRef]
- Blancato, V.S.; Magni, C.; Lolkema, J.S. Functional characterization and Me2+ ion specificity of a Ca2+-citrate transporter from Enterococcus faecalis. FEBS J. 2006, 273, 5121–5130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Manno, M.T.; Zuljan, F.; Alarcón, S.; Esteban, L.; Blancato, V.; Espariz, M.; Magni, C. Genetic and phenotypic features defining industrial relevant Lactococcus lactis, L. cremoris and L. lactis biovar. diacetylactis strains. J. Biotechnol. 2018, 282, 25–31. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Repizo, G.D.; Blancato, V.S.; Mortera, P.; Lolkema, J.S.; Magni, C. Biochemical and genetic characterization of the Enterococcus faecalis oxaloacetate decarboxylase complex. Appl. Environ. Microbiol. 2013, 79, 2882–2890. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sender, P.D.; Martín, M.G.; Peirú, S.; Magni, C. Characterization of an oxaloacetate decarboxylase that belongs to the malic enzyme family. FEBS Lett. 2004, 570, 217–222. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Espariz, M.; Repizo, G.; Blancato, V.; Mortera, P.; Alarcón, S.; Magni, C. Identification of malic and soluble oxaloacetate decarboxylase enzymes in Enterococcus faecalis. FEBS J. 2011, 278, 2140–2151. [Google Scholar] [CrossRef] [PubMed]
- Morishita, T.; Yajima, M. Incomplete operation of biosynthetic and bioenergetic functions of the citric acid cycle in multiple auxotrophic lactobacilli. Biosci. Biotechnol. Biochem. 1995, 59, 251–255. [Google Scholar] [CrossRef] [Green Version]
- Lapujade, P.; Cocaign-Bousquet, M.; Loubiere, P. Glutamate biosynthesis in Lactococcus lactis subsp. lactis NCDO 2118. Appl. Environ. Microbiol. 1998, 64, 2485–2489. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Baldwin, K.A.; O’Sullivan, D.J.; McKay, L.L. Identification of a gene cluster encoding Krebs cycle oxidative enzymes linked to the pyruvate carboxylase gene in Lactococcus lactis ssp. lactis C2. J. Dairy Sci. 2000, 83, 1912–1918. [Google Scholar] [CrossRef]
- Rea, M.C.; Cogan, T.M. Glucose prevents citrate metabolism by enterococci. Int. J. Food Microbiol. 2003, 88, 201–206. [Google Scholar] [CrossRef]
- Pudlik, A.M.; Lolkema, J.S. Citrate uptake in exchange with Intermediates in the citrate metabolic pathway in Lactococcus lactis IL1403. J. Bacteriol. 2011, 193, 706–714. [Google Scholar] [CrossRef] [Green Version]
- Schmitt, P.; Diviès, C.; Merlot, C. Utilization of citrate by Leuconostoc mesenteroides subsp. cremoris in continuous culture. Biotechnol. Lett. 1990, 12, 127–130. [Google Scholar] [CrossRef]
- Sarantinopoulos, P.; Kalantzopoulos, G.; Tsakalidou, E. Citrate metabolism by Enterococcus faecalis FAIR-E 229. Appl. Environ. Microbiol. 2001, 67, 5482–5487. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Starrenburg, M.J.; Hugenholtz, J. Citrate fermentation by Lactococcus and Leuconostoc spp. Appl. Environ. Microbiol. 1991, 57, 3535–3540. [Google Scholar] [CrossRef] [Green Version]
- García-Quintáns, N.; Magni, C.; de Mendoza, D.; López, P. The citrate transport system of Lactococcus lactis subsp. lactis biovar diacetylactis is induced by acid stress. Appl. Environ. Microbiol. 1998, 64, 850–857. [Google Scholar] [CrossRef] [Green Version]
- Marty-Teysset, C.; Posthuma, C.; Lolkema, J.S.; Schmitt, P.; Divies, C.; Konings, W.N. Proton motive force generation by citrolactic fermentation in Leuconostoc mesenteroides. J. Bacteriol. 1996, 178, 2178–2185. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martino, G.P.; Quintana, I.M.; Espariz, M.; Blancato, V.S.; Nizo, G.G.; Esteban, L.; Magni, C. Draft Genome sequences of four Enterococcus faecium strains isolated from Argentine cheese. Genome Announc. 2016, 4, 1515–1576. [Google Scholar] [CrossRef] [Green Version]
- D’Angelo, M.; Martino, G.P.; Blancato, V.S.; Espariz, M.; Hartke, A.; Sauvageot, N.; Benachour, A.; Alarcón, S.H.; Magni, C. Diversity of volatile organic compound production from leucine and citrate in Enterococcus faecium. Appl. Microbiol. Biotechnol. 2020, 104, 1175–1186. [Google Scholar] [CrossRef]
- Cselovszky, J.; Wolf, G.; Hammes, W.P. Production of formate, acetate, and succinate by anaerobic fermentation of Lactobacillus pentosus in the presence of citrate. Appl. Microbiol. Biotechnol. 1992, 37, 94–97. [Google Scholar] [CrossRef]
- Dudley, E.G.; Steele, J.L. Succinate production and citrate catabolism by Cheddar cheese nonstarter lactobacilli. J. Appl. Microbiol. 2005, 98, 14–23. [Google Scholar] [CrossRef]
- Larrouture-Thiveyrat, C.; Montel, M.-C. Effects of environmental factors on leucine catabolism by Carnobacterium piscicola. Int. J. Food Microbiol. 2003, 81, 177–184. [Google Scholar] [CrossRef]
- Afzal, M.I.; Delaunay, S.; Paris, C.; Borges, F.; Revol-Junelles, A.-M.; Cailliez-Grimal, C. Identification of metabolic pathways involved in the biosynthesis of flavor compound 3-methylbutanal from leucine catabolism by Carnobacterium maltaromaticum LMA 28. Int. J. Food Microbiol. 2012, 157, 332–339. [Google Scholar] [CrossRef] [PubMed]
- Afzal, M.I.; Boulahya, K.-A.; Paris, C.; Delaunay, S.; Cailliez-Grimal, C. Effect of oxygen on the biosynthesis of flavor compound 3-methylbutanal from leucine catabolism during batch culture in Carnobacterium maltaromaticum LMA 28. J. Dairy Sci. 2013, 96, 352–359. [Google Scholar] [CrossRef] [PubMed]


| Strain | Isolated from | Citrate Metabolic Phenotype a | Genes Encoding Specific Pathway | Ref. | |
|---|---|---|---|---|---|
| Pathway 1 b | Pathway 2 c | ||||
| Cm 4-1d | Sphagnum pond | − | + | + | [13] |
| Cm 6-1 | Sphagnum pond | (+) | + | + | [13] |
| Cm 1-2 | Sphagnum pond | − | − | + | [13] |
| Cm 3-1 | Sphagnum pond | (+)(d) | − | + | [13] |
| Cm 5-1 | Sphagnum pond | (+)(d) | − | + | [13] |
| ATCC35586 | Diseased salmon | (+) | − | + | [6] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, H.; Ramia, N.E.; Borges, F.; Revol-Junelles, A.-M.; Vogensen, F.K.; Leisner, J.J. Identification of Potential Citrate Metabolism Pathways in Carnobacterium maltaromaticum. Microorganisms 2021, 9, 2169. https://doi.org/10.3390/microorganisms9102169
Li H, Ramia NE, Borges F, Revol-Junelles A-M, Vogensen FK, Leisner JJ. Identification of Potential Citrate Metabolism Pathways in Carnobacterium maltaromaticum. Microorganisms. 2021; 9(10):2169. https://doi.org/10.3390/microorganisms9102169
Chicago/Turabian StyleLi, Heng, Nancy E. Ramia, Frédéric Borges, Anne-Marie Revol-Junelles, Finn Kvist Vogensen, and Jørgen J. Leisner. 2021. "Identification of Potential Citrate Metabolism Pathways in Carnobacterium maltaromaticum" Microorganisms 9, no. 10: 2169. https://doi.org/10.3390/microorganisms9102169
APA StyleLi, H., Ramia, N. E., Borges, F., Revol-Junelles, A.-M., Vogensen, F. K., & Leisner, J. J. (2021). Identification of Potential Citrate Metabolism Pathways in Carnobacterium maltaromaticum. Microorganisms, 9(10), 2169. https://doi.org/10.3390/microorganisms9102169






