Characterization of the Microbiome of Corals with Stony Coral Tissue Loss Disease along Florida’s Coral Reef
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Sample Processing
2.3. Data Analysis
3. Results
3.1. Microbial Community Analysis
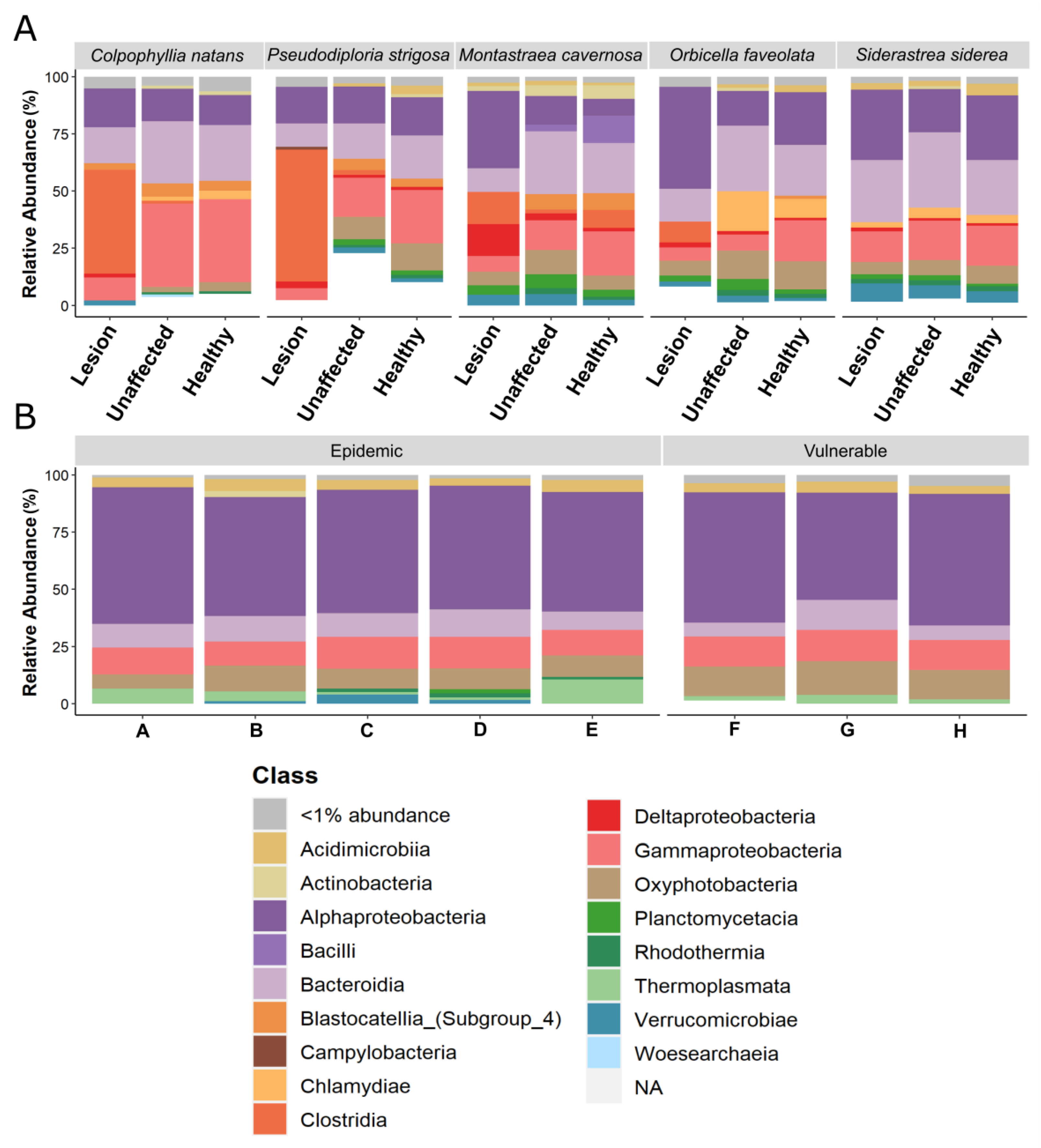
3.1.1. Microbial Communities among Sample Types
3.1.2. Microbial Communities among Sites within Zones
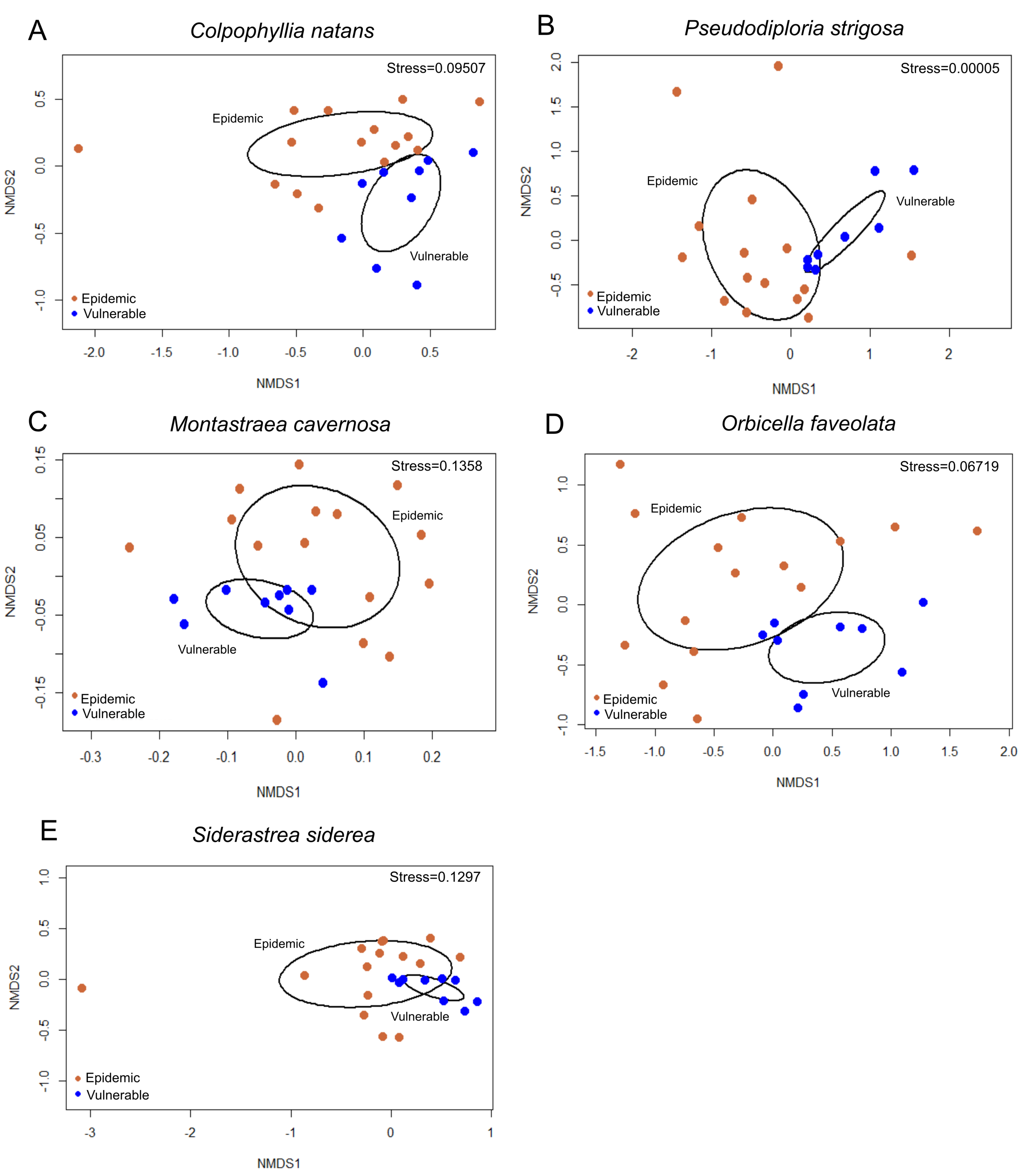
3.2. Beta Diversity Analysis
3.2.1. Beta Diversity between Vulnerable and Epidemic Zones
3.2.2. Beta Diversity among Sites within Zones
3.2.3. Beta Diversity among Coral Tissue Sample Types in the Epidemic Zone
3.2.4. Beta Diversity of Apparently Healthy Colony Tissue in Vulnerable and Epidemic Zones
3.3. Alpha Diversity Analysis
3.3.1. Alpha Diversity between Vulnerable and Epidemic Zones
3.3.2. Alpha Diversity among Sites within Zones
3.3.3. Alpha Diversity among Coral Tissue Sample Types in the Epidemic Zone
3.3.4. Alpha Diversity of Apparently Healthy Colony Tissue in Vulnerable and Epidemic Zones
3.4. Relative Abundance Analysis of Bacterial Class
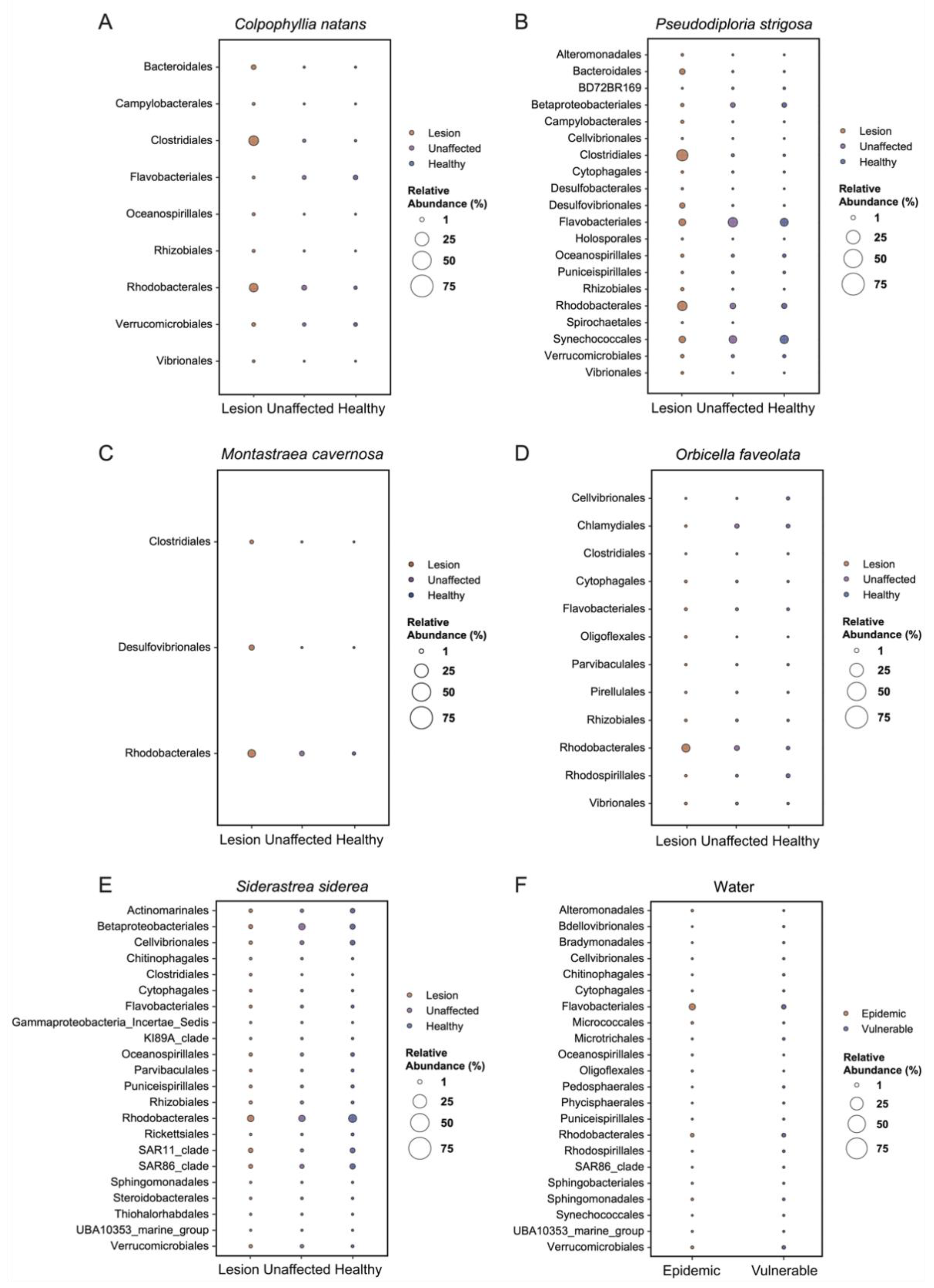
3.5. Analysis of Composition of Microbiomes
4. Discussion
4.1. Diversity Indices
4.2. Differences Were Detected among Sites and Tissue Sample Types
4.3. Rhodobacterales and Clostridiales in SCTLD Lesions
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rohwer, F.; Seguritan, V.; Azam, F.; Knowlton, N. Diversity and Distribution of Coral-Associated Bacteria. Mar. Ecol. Prog. Ser. 2002, 243, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Wegley, L.; Yu, Y.; Breitbart, M.; Casas, V.; Kline, D.; Rohwer, F. Coral-Associated Archaea. Mar. Ecol. Prog. Ser. 2004, 273, 89–96. [Google Scholar] [CrossRef]
- Bourne, D.G.; Morrow, K.M.; Webster, N.S. Insights into the Coral Microbiome: Underpinning the Health and Resilience of Reef Ecosystems. Annu. Rev. Microbiol. 2016, 70, 317–340. [Google Scholar] [CrossRef]
- Ainsworth, T.D.; Fordyce, A.J.; Camp, E.F. The Other Microeukaryotes of the Coral Reef Microbiome. Trends Microbiol. 2017, 25, 980–991. [Google Scholar] [CrossRef] [PubMed]
- Clerissi, C.; Brunet, S.; Vidal-Dupiol, J.; Adjeroud, M.; Lepage, P.; Guillou, L.; Escoubas, J.-M.; Toulza, E. Protists Within Corals: The Hidden Diversity. Front. Microbiol. 2018, 9, 2043. [Google Scholar] [CrossRef] [PubMed]
- Rädecker, N.; Pogoreutz, C.; Voolstra, C.R.; Wiedenmann, J.; Wild, C. Nitrogen Cycling in Corals: The Key to Understanding Holobiont Functioning? Trends Microbiol. 2015, 23, 490–497. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Benavides, M.; Bednarz, V.N.; Ferrier-Pagès, C. Diazotrophs: Overlooked Key Players within the Coral Symbiosis and Tropical Reef Ecosystems? Front. Mar. Sci. 2017, 4, 10. [Google Scholar] [CrossRef] [Green Version]
- Vanwonterghem, I.; Webster, N.S. Coral Reef Microorganisms in a Changing Climate. iScience 2020, 23, 100972. [Google Scholar] [CrossRef]
- Matthews, J.L.; Raina, J.; Kahlke, T.; Seymour, J.R.; Oppen, M.J.H.; Suggett, D.J. Symbiodiniaceae-bacteria Interactions: Rethinking Metabolite Exchange in Reef-building Corals as Multi-partner Metabolic Networks. Env. Microbiol. 2020, 22, 1675–1687. [Google Scholar] [CrossRef] [Green Version]
- Ritchie, K. Regulation of Microbial Populations by Coral Surface Mucus and Mucus-Associated Bacteria. Mar. Ecol. Prog. Ser. 2006, 322, 1–14. [Google Scholar] [CrossRef]
- Peixoto, R.S.; Rosado, P.M.; de Assis Leite, D.C.; Rosado, A.S.; Bourne, D.G. Beneficial Microorganisms for Corals (BMC): Proposed Mechanisms for Coral Health and Resilience. Front. Microbiol. 2017, 8, 341. [Google Scholar] [CrossRef] [Green Version]
- Thompson, J.R.; Rivera, H.E.; Closek, C.J.; Medina, M. Microbes in the Coral Holobiont: Partners through Evolution, Development, and Ecological Interactions. Front. Cell. Infect. Microbiol. 2015, 4, 176. [Google Scholar] [CrossRef]
- Littman, R.; Willis, B.L.; Bourne, D.G. Metagenomic Analysis of the Coral Holobiont during a Natural Bleaching Event on the Great Barrier Reef. Environ. Microbiol. Rep. 2011, 3, 651–660. [Google Scholar] [CrossRef]
- van Oppen, M.J.H.; Blackall, L.L. Coral Microbiome Dynamics, Functions and Design in a Changing World. Nat. Rev. Microbiol. 2019, 17, 557–567. [Google Scholar] [CrossRef] [PubMed]
- Boilard, A.; Dubé, C.E.; Gruet, C.; Mercière, A.; Hernandez-Agreda, A.; Derome, N. Defining Coral Bleaching as a Microbial Dysbiosis within the Coral Holobiont. Microorganisms 2020, 8, 1682. [Google Scholar] [CrossRef]
- Hoegh-Guldberg, O.; Mumby, P.J.; Hooten, A.J.; Steneck, R.S.; Greenfield, P.; Gomez, E.; Harvell, C.D.; Sale, P.F.; Edwards, A.J.; Caldeira, K.; et al. Coral Reefs under Rapid Climate Change and Ocean Acidification. Science 2007, 14, 1737–1742. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hughes, T.P.; Barnes, M.L.; Bellwood, D.R.; Cinner, J.E.; Cumming, G.S.; Jackson, J.B.C.; Kleypas, J.; van de Leemput, I.A.; Lough, J.M.; Morrison, T.H.; et al. Coral Reefs in the Anthropocene. Nature 2017, 546, 82–90. [Google Scholar] [CrossRef] [PubMed]
- Epstein, P.R.; Sherman, K.; Spanger-Siegfried, E.; Langston, A.; Prasad, S.; McKay, B. Marine Ecosystems: Emerging Diseases as Indicators of Change. In Health Ecological and Economic Dimensions (HEED), NOAA Global Change Program; Center for Health and the Global Environment: Boston, MA, USA, 1998; 85p. [Google Scholar]
- Green, E.P.; Bruckner, A.W. The Significance of Coral Disease Epizootiology for Coral Reef Conservation. Biol. Conserv. 2000, 96, 347–361. [Google Scholar] [CrossRef]
- Weil, E.; Urreiztieta, I.; Garzôn-Ferreira, J. Geographic Variability in the Incidence of Coral and Octocoral Disease in the Wider Caribbean. In Proceedings of the 9th International Coral Reef Symposium, Bali, Indonesia, 23–27 October 2000; Volume 2, pp. 1231–1238. [Google Scholar]
- Willis, B.L.; Page, C.A.; Dinsdale, E.A. Coral Disease on the Great Barrier Reef. In Coral Health and Disease; Rosenberg, E., Loya, Y., Eds.; Springer: Berlin/Heidelberg, Germany, 2004; pp. 69–104. [Google Scholar] [CrossRef]
- Pollock, F.J.; Lamb, J.B.; Field, S.N.; Heron, S.F.; Schaffelke, B.; Shedrawi, G.; Bourne, D.G.; Willis, B.L. Sediment and Turbidity Associated with Offshore Dredging Increase Coral Disease Prevalence on Nearby Reefs. PLoS ONE 2014, 9, e102498. [Google Scholar] [CrossRef] [Green Version]
- Precht, W.F.; Gintert, B.E.; Robbart, M.L.; Fura, R.; van Woesik, R. Unprecedented Disease-Related Coral Mortality in Southeastern Florida. Sci. Rep. 2016, 6, 31374. [Google Scholar] [CrossRef]
- Atlantic and Gulf Rapid Reef Assessment. Available online: https://www.agrra.org/coral-disease-outbreak/ (accessed on 7 July 2021).
- Florida Department of Environmental Protection. Available online: https://floridadep.gov/sites/default/files/Copy%20of%20StonyCoralTissueLossDisease_CaseDefinition%20final%2010022018.pdf (accessed on 7 July 2021).
- Aeby, G.S.; Ushijima, B.; Campbell, J.E.; Jones, S.; Williams, G.J.; Meyer, J.L.; Häse, C.; Paul, V.J. Pathogenesis of a Tissue Loss Disease Affecting Multiple Species of Corals Along the Florida Reef Tract. Front. Mar. Sci. 2019, 6, 678. [Google Scholar] [CrossRef]
- Estrada-Saldívar, N.; Molina-Hernández, A.; Pérez-Cervantes, E.; Medellín-Maldonado, F.; González-Barrios, F.J.; Alvarez-Filip, L. Reef-Scale Impacts of the Stony Coral Tissue Loss Disease Outbreak. Coral Reefs 2020, 39, 861–866. [Google Scholar] [CrossRef]
- Estrada-Saldívar, N.; Quiroga-García, B.A.; Pérez-Cervantes, E.; Rivera-Garibay, O.O.; Alvarez-Filip, L. Effects of the Stony Coral Tissue Loss Disease Outbreak on Coral Communities and the Benthic Composition of Cozumel Reefs. Front. Mar. Sci. 2021, 8, 632777. [Google Scholar] [CrossRef]
- Neely, K.L.; Macaulay, K.A.; Hower, E.K.; Dobler, M.A. Effectiveness of Topical Antibiotics in Treating Corals Affected by Stony Coral Tissue Loss Disease. PeerJ 2020, 8, e9289. [Google Scholar] [CrossRef] [PubMed]
- Landsberg, J.H.; Kiryu, Y.; Peters, E.C.; Wilson, P.W.; Perry, N.; Waters, Y.; Maxwell, K.E.; Huebner, L.K.; Work, T.M. Stony Coral Tissue Loss Disease in Florida Is Associated With Disruption of Host–Zooxanthellae Physiology. Front. Mar. Sci. 2020, 7, 576013. [Google Scholar] [CrossRef]
- Dobbelaere, T.; Muller, E.M.; Gramer, L.J.; Holstein, D.M.; Hanert, E. Coupled Epidemio-Hydrodynamic Modeling to Understand the Spread of a Deadly Coral Disease in Florida. Front. Mar. Sci. 2020, 7, 591881. [Google Scholar] [CrossRef]
- Muller, E.M.; Sartor, C.; Alcaraz, N.I.; van Woesik, R. Spatial Epidemiology of the Stony-Coral-Tissue-Loss Disease in Florida. Front. Mar. Sci. 2020, 7, 163. [Google Scholar] [CrossRef] [Green Version]
- Dahlgren, C.; Pizarro, V.; Sherman, K.; Greene, W.; Oliver, J. Spatial and Temporal Patterns of Stony Coral Tissue Loss Disease Outbreaks in The Bahamas. Front. Mar. Sci. 2021, 8, 682114. [Google Scholar] [CrossRef]
- Williams, S.D.; Walter, C.S.; Muller, E.M. Fine Scale Temporal and Spatial Dynamics of the Stony Coral Tissue Loss Disease Outbreak Within the Lower Florida Keys. Front. Mar. Sci. 2021, 8, 631776. [Google Scholar] [CrossRef]
- Ushijima, B.; Meyer, J.L.; Thompson, S.; Pitts, K.; Marusich, M.F.; Tittl, J.; Weatherup, E.; Reu, J.; Wetzell, R.; Aeby, G.S.; et al. Disease Diagnostics and Potential Coinfections by Vibrio coralliilyticus During an Ongoing Coral Disease Outbreak in Florida. Front. Microbiol. 2020, 11, 569354. [Google Scholar] [CrossRef]
- Walker, B.K.; Turner, N.R.; Noren, H.K.G.; Buckley, S.F.; Pitts, K.A. Optimizing Stony Coral Tissue Loss Disease (SCTLD) Intervention Treatments on Montastraea cavernosa in an Endemic Zone. Front. Mar. Sci. 2021, 8, 666224. [Google Scholar] [CrossRef]
- Neely, K.L.; Shea, C.P.; Macaulay, K.A.; Hower, E.K.; Dobler, M.A. Short- and Long-Term Effectiveness of Coral Disease Treatments. Front. Mar. Sci. 2021, 8, 675349. [Google Scholar] [CrossRef]
- Walton, C.J.; Hayes, N.K.; Gilliam, D.S. Impacts of a Regional, Multi-Year, Multi-Species Coral Disease Outbreak in Southeast Florida. Front. Mar. Sci. 2018, 5, 323. [Google Scholar] [CrossRef]
- Meyer, J.L.; Castellanos-Gell, J.; Aeby, G.S.; Häse, C.C.; Ushijima, B.; Paul, V.J. Microbial Community Shifts Associated With the Ongoing Stony Coral Tissue Loss Disease Outbreak on the Florida Reef Tract. Front. Microbiol. 2019, 10, 2244. [Google Scholar] [CrossRef] [PubMed]
- Rosales, S.M.; Clark, A.S.; Huebner, L.K.; Ruzicka, R.R.; Muller, E.M. Rhodobacterales and Rhizobiales Are Associated With Stony Coral Tissue Loss Disease and Its Suspected Sources of Transmission. Front. Microbiol. 2020, 11, 681. [Google Scholar] [CrossRef] [PubMed]
- Work, T.M. Final Report on Electron Microscopy of Florida Corals Affected with Stony Coral Tissue Loss Disease (SCTLD). U.S. Geological Survey Diagnostic Case Report 2021. Available online: https://www.caymancompass.com/wp-content/uploads/2021/08/USGS-SCTLD-research-report.pdf?__cf_chl_captcha_tk__=pmd_h_fJ47Hee45Ys_Bx2Wealj_1yVhP3i3zSz3My_PvQ_Y-1634709587-0-gqNtZGzNAyWjcnBszQil (accessed on 31 August 2021).
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Lozupone, C.A.; Turnbaugh, P.J.; Fierer, N.; Knight, R. Global Patterns of 16S rRNA Diversity at a Depth of Millions of Sequences per Sample. Proc. Natl. Acad. Sci. USA 2011, 108 (Suppl. 1), 4516–4522. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takai, K.; Horikoshi, K. Rapid Detection and Quantification of Members of the Archaeal Community by Quantitative PCR using Fluorogenic Probes. Appl Environ. Microbiol. 2000, 66, 5066–5072. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- R Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing. Available online: https://www.R-project.org/ (accessed on 29 August 2021).
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-Resolution Sample Inference from Illumina Amplicon Data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Callahan, B. Silva Taxonomic Training Data Formatted for DADA2 (Silva Version 132). Zenodo 2018. [Google Scholar] [CrossRef]
- McMurdie, P.J.; Holmes, S. phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oksanen, J.; Blanchet, F.G.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; et al. Vegan: Community Ecology Package. R Package Version 2.5-7. 2020. Available online: https://CRAN.R-project.org/package=vegan (accessed on 16 October 2021).
- Martinez, A. PairwiseAdonis: Pairwise Multilevel Comparison Using Adonis R Package Version 0.3. 2019. Available online: https://github.com/pmartinezarbizu/pairwiseAdonis (accessed on 16 October 2021).
- Mandal, S.; Van Treuren, W.; White, R.A.; Eggesbø, M.; Knight, R.; Peddada, S.D. Analysis of Composition of Microbiomes: A Novel Method for Studying Microbial Composition. Microb. Ecol. Health Dis. 2015, 26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iwanowicz, D.D.; Schill, W.B.; Woodley, C.M.; Bruckner, A.; Neely, K.; Briggs, K.M. Exploring the Stony Coral Tissue Loss Disease Bacterial Pathobiome. Microbiology 2020. preprint. [Google Scholar] [CrossRef]
- Becker, C.C.; Brandt, M.; Miller, C.A.; Apprill, A. Stony Coral Tissue Loss Disease Biomarker Bacteria Identified in Corals and Overlying Waters Using a Rapid Field-Based Sequencing Approach; preprint. Microbiology 2021. [Google Scholar] [CrossRef]
- Zaneveld, J.R.; Burkepile, D.E.; Shantz, A.A.; Pritchard, C.E.; McMinds, R.; Payet, J.P.; Welsh, R.; Correa, A.M.S.; Lemoine, N.P.; Rosales, S.; et al. Overfishing and Nutrient Pollution Interact with Temperature to Disrupt Coral Reefs down to Microbial Scales. Nat. Commun. 2016, 7, 11833. [Google Scholar] [CrossRef]
- Weber, L.; Gonzalez-Díaz, P.; Armenteros, M.; Apprill, A. The Coral Ecosphere: A Unique Coral Reef Habitat That Fosters Coral–Microbial Interactions. Limnol. Oceanogr. 2019, 64, 2373–2388. [Google Scholar] [CrossRef] [Green Version]
- Eaton, K.R.; Landsberg, J.H.; Kiryu, Y.; Peters, E.C.; Muller, E.M. Measuring Stony Coral Tissue Loss Disease Induction and Lesion Progression Within Two Intermediately Susceptible Species, Montastraea cavernosa and Orbicella faveolata. Front. Mar. Sci. 2021, 8, 717265. [Google Scholar] [CrossRef]
- Zaneveld, J.; McMinds, R.; Vega Thurber, R. Stress and stability: Applying the Anna Karenina principle to animal microbiomes. Nat. Microbiol. 2017, 2, 17121. [Google Scholar] [CrossRef]
- Rosales, S.M.; Miller, M.W.; Williams, D.E.; Traylor-Knowles, N.; Young, B.; Serrano, X.M. Microbiome Differences in Disease-Resistant vs. Susceptible Acropora Corals Subjected to Disease Challenge Assays. Sci. Rep. 2019, 9, 18279. [Google Scholar] [CrossRef] [Green Version]
- MacKnight, N.J.; Cobleigh, K.; Lasseigne, D.; Chaves-Fonnegra, A.; Gutting, A.; Dimos, B.; Antoine, J.; Fuess, L.; Ricci, C.; Butler, C.; et al. Microbial Dysbiosis Reflects Disease Resistance in Diverse Coral Species. Commun. Biol. 2021, 4, 679. [Google Scholar] [CrossRef] [PubMed]
- McDevitt-Irwin, J.M.; Baum, J.K.; Garren, M.; Vega Thurber, R.L. Responses of Coral-Associated Bacterial Communities to Local and Global Stressors. Front. Mar. Sci. 2017, 4, 262. [Google Scholar] [CrossRef] [Green Version]
- Vega Thurber, R.; Willner-Hall, D.; Rodriguez-Mueller, B.; Desnues, C.; Edwards, R.A.; Angly, F.; Dinsdale, E.; Kelly, L.; Rohwer, F. Metagenomic Analysis of Stressed Coral Holobionts. Environ. Microbiol. 2009, 11. [Google Scholar] [CrossRef]
- Glasl, B.; Herndl, G.J.; Frade, P.R. The Microbiome of Coral Surface Mucus Has a Key Role in Mediating Holobiont Health and Survival upon Disturbance. ISME J. 2016, 10, 2280–2292. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gignoux-Wolfsohn, S.A.; Aronson, F.M.; Vollmer, S.V. Complex Interactions between Potentially Pathogenic, Opportunistic, and Resident Bacteria Emerge during Infection on a Reef-Building Coral. FEMS Microbiol. Ecol. 2017, 93, fix080. [Google Scholar] [CrossRef] [Green Version]
- Sharp, W.C.; Shea, C.P.; Maxwell, K.E.; Muller, E.M.; Hunt, J.H. Evaluating the Small-Scale Epidemiology of the Stony-Coral -Tissue-Loss-Disease in the Middle Florida Keys. PLoS ONE 2020, 15, e0241871. [Google Scholar] [CrossRef]
- Cooney, R.P.; Pantos, O.; Le Tissier, M.D.A.; Barer, M.R.; O’Donnell, A.G.; Bythell, J.C. Characterization of the Bacterial Consortium Associated with Black Band Disease in Coral using Molecular Microbiological Techniques. Environ. Microbiol. 2002, 4, 401–413. [Google Scholar] [CrossRef]
- Pantos, O.; Cooney, R.P.; Le Tissier, M.D.A.; Barer, M.R.; O’Donnell, A.G.; Bythell, J.C. The Bacterial Ecology of a Plague-Like Disease Affecting the Caribbean Coral Montastrea Annularis. Environ. Microbiol. 2003, 5, 370–382. [Google Scholar] [CrossRef]
- Pantos, O.; Bythell, J.C. Bacterial Community Structure Associated with White Band Disease in the Elkhorn Coral Acropora palmata Determined using Culture-Independent 16S rRNA Techniques. Dis. Aqu. Org. 2006, 69, 79–88. [Google Scholar] [CrossRef]
- Sunagawa, S.; DeSantis, T.Z.; Piceno, Y.M.; Brodie, E.L.; DeSalvo, M.K.; Voolstra, C.R.; Weil, E.; Andersen, G.L.; Medina, M. Bacterial Diversity and White Plague Disease-Associated Community Changes in the Caribbean Coral Montastraea faveolata. ISME J. 2009, 3, 512–521. [Google Scholar] [CrossRef] [Green Version]
- Mouchka, M.E.; Hewson, I.; Harvell, C.D. Coral-Associated Bacterial Assemblages: Current Knowledge and the Potential for Climate-Driven Impacts. Integr. Comp. Biol. 2010, 50, 662–674. [Google Scholar] [CrossRef] [PubMed]
- Soffer, N.; Zaneveld, J.; Vega Thurber, R. Phage-Bacteria Network Analysis and Its Implication for the Understanding of Coral Disease: Phage-Bacteria Interaction Networks of Corals. Environ. Microbiol. 2015, 17, 1203–1218. [Google Scholar] [CrossRef] [PubMed]
- Dang, H.; Lovell, C.R. Bacterial Primary Colonization and Early Succession on Surfaces in Marine Waters as Determined by Amplified rRNA Gene Restriction Analysis and Sequence Analysis of 16S rRNA Genes. Appl. Environ. Microbiol. 2000, 66, 467–475. [Google Scholar] [CrossRef] [Green Version]
- Dang, H.; Li, T.; Chen, M.; Huang, G. Cross-Ocean Distribution of Rhodobacterales Bacteria as Primary Surface Colonizers in Temperate Coastal Marine Waters. Appl. Environ. Microbiol. 2008, 74, 52–60. [Google Scholar] [CrossRef] [Green Version]
- Mayali, X.; Weber, P.K.; Mabery, S.; Pett-Ridge, J. Phylogenetic Patterns in the Microbial Response to Resource Availability: Amino Acid Incorporation in San Francisco Bay. PLoS ONE 2014, 9, e95842. [Google Scholar] [CrossRef] [PubMed]
- Brinkhoff, T.; Bach, G.; Heidorn, T.; Liang, L.; Schlingloff, A.; Simon, M. Antibiotic Production by a Roseobacter Clade-Affiliated Species from the German Wadden Sea and Its Antagonistic Effects on Indigenous Isolates. Appl. Environ. Microbiol. 2004, 70, 2560–2565. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burge, C.A.; Eakin, C.M.; Friedman, C.S.; Froelich, B.; Hershberger, P.K.; Hofmann, E.E.; Petes, L.E.; Prager, K.C.; Weil, E.; Willis, B.L.; et al. Climate Change Influences on Marine Infectious Diseases: Implications for Management and Society. Mar. Sci. 2013, 6, 249–277. [Google Scholar] [CrossRef] [Green Version]
- Sweet, M.J.; Bythell, J.C.; Nugues, M.M. Algae as Reservoirs for Coral Pathogens. PLoS ONE 2013, 8, e69717. [Google Scholar] [CrossRef] [Green Version]
- Leiblein, M.; Wagner, N.; Adam, E.H.; Frank, J.; Marzi, I.; Nau, C. Clostridial Gas Gangrene - A Rare but Deadly Infection: Case Series and Comparison to Other Necrotizing Soft Tissue Infections. Orthop. Surg. 2020, 12, 1733–1747. [Google Scholar] [CrossRef] [PubMed]
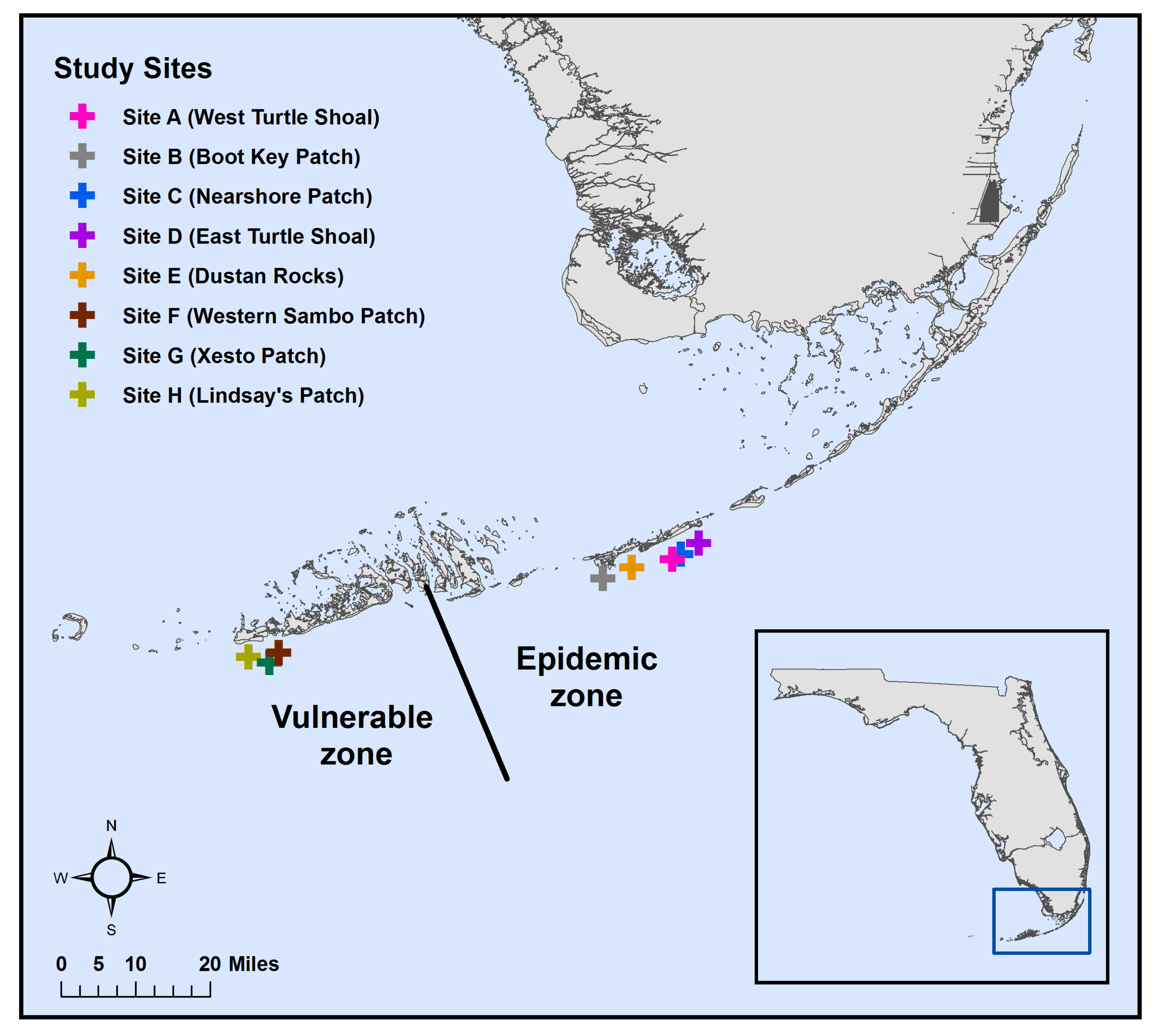




| Date | Zone | Site Letter Code 1 | Coral Species | Lesion/ Site | Unaffected/ Site | Healthy/ Site | Water/ Site |
|---|---|---|---|---|---|---|---|
| 04/09–04/27/18 | Epidemic | A–E | CNAT | 5 | 5 | 3 | - |
| 04/09–04/27/18 | Epidemic | A–E | PSTR | 4–5 | 4–5 | 3 | - |
| 04/09–04/27/18 | Epidemic | A–E | MCAV | 5 | 5 | 3 | - |
| 04/09–04/27/18 | Epidemic | A–E | OFAV | 3–5 | 3–5 | 3 | - |
| 04/09–04/27/18 | Epidemic | A–E | SSID | 5 | 5 | 3 | - |
| 04/09–04/27/18 | Epidemic | A–E | - | - | - | - | 3 |
| 05/08–06/05/18 | Vulnerable | F–H | CNAT | - | - | 3 | - |
| 05/08–06/05/18 | Vulnerable | F–H | PSTR | - | - | 2–3 | - |
| 05/08–06/05/18 | Vulnerable | F–H | MCAV | - | - | 3 | - |
| 05/08–06/05/18 | Vulnerable | F–H | OFAV | - | - | 3 | - |
| 05/08–06/05/18 | Vulnerable | F–H | SSID | - | - | 3 | - |
| 05/08–06/05/18 | Vulnerable | F–H | - | - | - | - | 3 |
| Total | 122 | 122 | 119 | 24 |
| ASV ID | Family, Genus | Reference |
|---|---|---|
| 6 | Rhodobacteraceae, Ruegeria | [39,51] |
| 21 | Rhodobacteraceae, Thalassobius | [39,40,52] |
| 22 | Rhodobacteraceae, Shimia | [39,40,51,52] |
| 50 | Peptostreptococcaceae, Tepidibacter | [39,51] |
| 57 | Rhodobacteraceae, Nautella | [39] |
| 116 | Rhodobacteraceae, Planktotalea | [39,51] |
| 124 | Rhodobacteraceae, Salinihabitans | [39] |
| 148 | Rhodobacteraceae, unclassified | [39] |
| 208 | Rhodobacteraceae, Nioella | [39,51] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Clark, A.S.; Williams, S.D.; Maxwell, K.; Rosales, S.M.; Huebner, L.K.; Landsberg, J.H.; Hunt, J.H.; Muller, E.M. Characterization of the Microbiome of Corals with Stony Coral Tissue Loss Disease along Florida’s Coral Reef. Microorganisms 2021, 9, 2181. https://doi.org/10.3390/microorganisms9112181
Clark AS, Williams SD, Maxwell K, Rosales SM, Huebner LK, Landsberg JH, Hunt JH, Muller EM. Characterization of the Microbiome of Corals with Stony Coral Tissue Loss Disease along Florida’s Coral Reef. Microorganisms. 2021; 9(11):2181. https://doi.org/10.3390/microorganisms9112181
Chicago/Turabian StyleClark, Abigail S., Sara D. Williams, Kerry Maxwell, Stephanie M. Rosales, Lindsay K. Huebner, Jan H. Landsberg, John H. Hunt, and Erinn M. Muller. 2021. "Characterization of the Microbiome of Corals with Stony Coral Tissue Loss Disease along Florida’s Coral Reef" Microorganisms 9, no. 11: 2181. https://doi.org/10.3390/microorganisms9112181






