Genomic Comparison of Conjugative Plasmids from Salmonella enterica and Escherichia coli Encoding Beta-Lactamases and Capable of Mobilizing Kanamycin Resistance Col-like Plasmids
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strains
2.2. KanR Plasmid Mobilization Tests on Agar
2.3. Sequencing
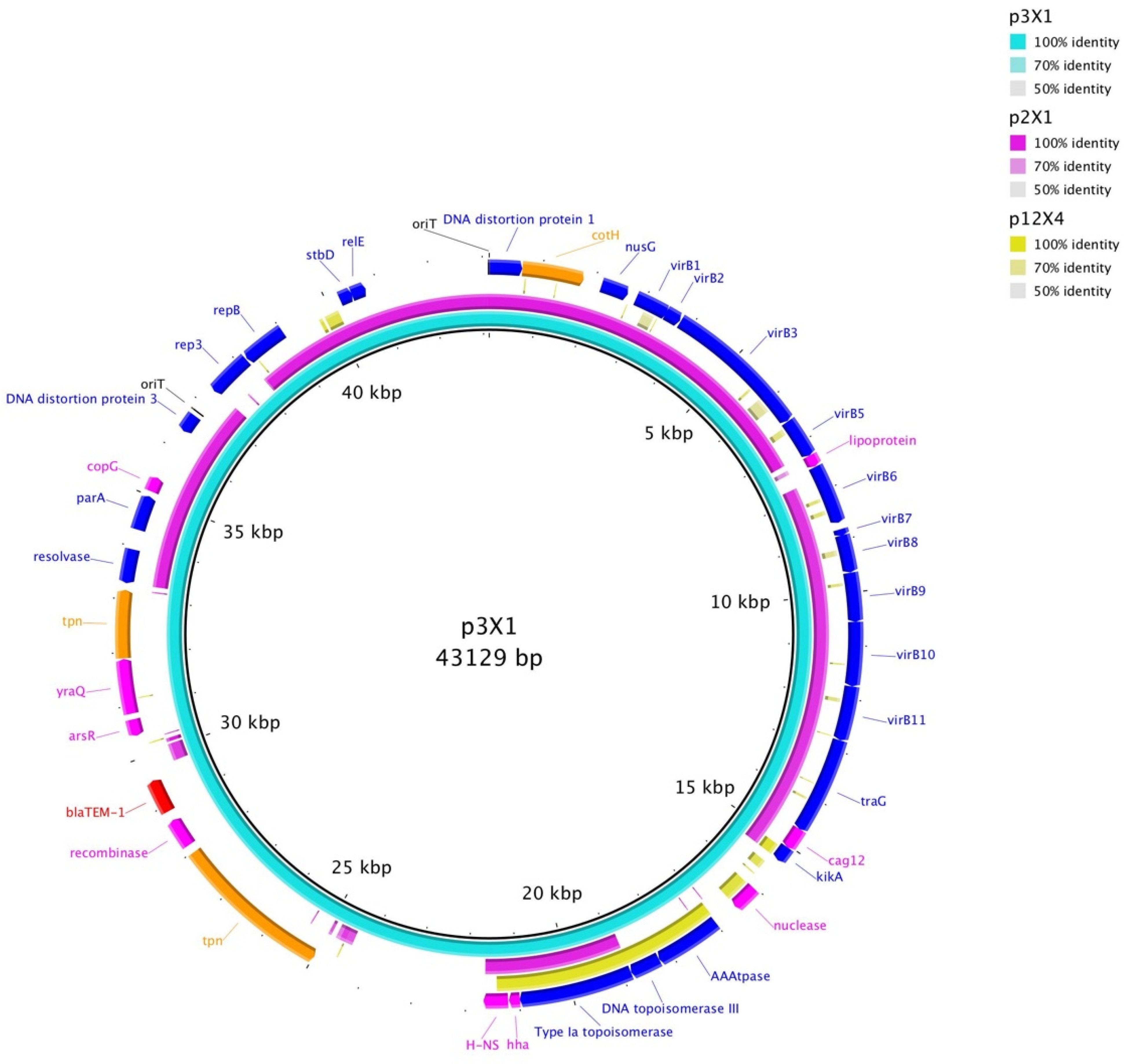
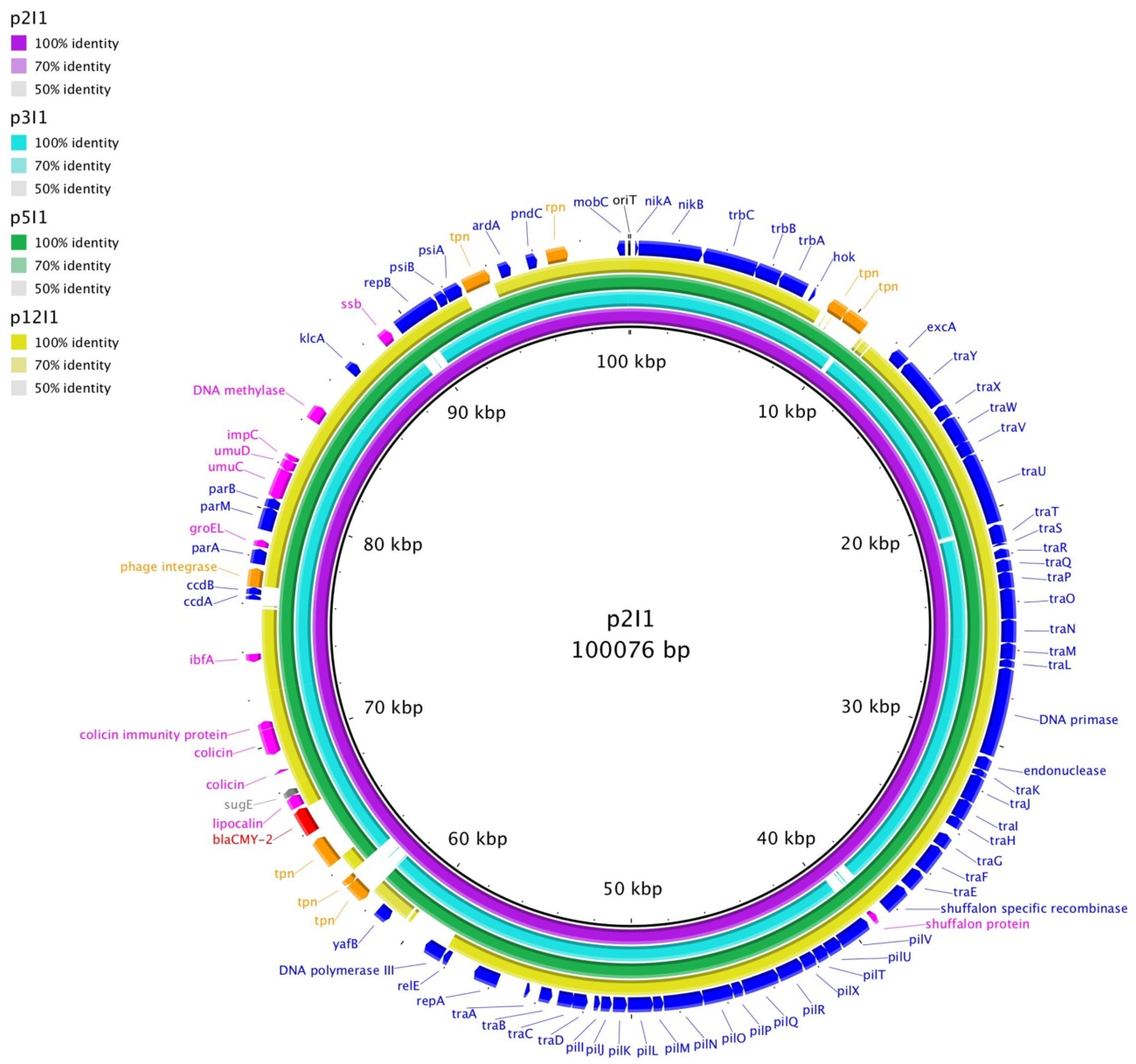
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Disclaimer
References
- Scallan, E.; Hoekstra, R.M.; Angulo, F.J.; Tauxe, R.V.; Widdowson, M.A.; Roy, S.L.; Jones, J.L.; Griffin, P.M. Foodborne illness acquired in the United States—Major pathogens. Emerg. Infect. Dis. 2011, 17, 7–15. [Google Scholar] [CrossRef]
- Tack, D.M.; Ray, L.; Griffin, P.M.; Cieslak, P.R.; Dunn, J.; Rissman, T.; Jervis, R.; Lathrop, S.; Muse, A.; Duwell, M.; et al. Preliminary incidence and trends of infections with pathogenes transmitted commonly through food—Foodborne diseases active surveillance network, 10 U.S. sites, 2016–2019. Morb. Mortal. Wkly. Rep. 2020, 69, 509–514. [Google Scholar] [CrossRef] [PubMed]
- CDC. Antibiotic Resistance Threats in the United States; CDC: Atlanta, GA, USA, 2019. [Google Scholar]
- Carattoli, A. Plasmids and the spread of resistance. Int. J. Med. Microbiol. 2013, 303, 298–304. [Google Scholar] [CrossRef]
- Folster, J.P.; Pecic, G.; McCullough, A.; Rickert, R.; Whichard, J.M. Characterization of bla(CMY)-encoding plasmids among Salmonella isolated in the United States in 2007. Foodborne Pathog. Dis. 2011, 8, 1289–1294. [Google Scholar] [CrossRef] [PubMed]
- Loftie-Eaton, W.; Rawlings, D.E. Diversity, biology and evolution of IncQ-family plasmids. Plasmid 2012, 67, 15–34. [Google Scholar] [CrossRef] [PubMed]
- Zechner, E.L.; Moncalián, G.; de la Cruz, F. Relaxases and Plasmid Transfer in Gram-Negative Bacteria. In Type IV Secretion in Gram-Negative and Gram-Positive Bacteria; Backert, S., Grohmann, E., Eds.; Springer International Publishing: Cham, Switzerland, 2017; pp. 93–113. [Google Scholar] [CrossRef]
- Francia, M.V.; Varsaki, A.; Garcillan-Barcia, M.P.; Latorre, A.; Drainas, C.; de la Cruz, F. A classification scheme for mobilization regions of bacterial plasmids. FEMS Microbiol. Rev. 2004, 28, 79–100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Warren, G.J.; Saul, M.W.; Sherratt, D.J. ColE1 plasmid mobility: Essential and conditional functions. Mol. Gen. Genet. 1979, 170, 103–107. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.Y.; Nace, G.W.; Solow, B.; Fratamico, P. Complete nucleotide sequences of 84.5- and 3.2-kb plasmids in the multi-antibiotic resistant Salmonella enterica serovar Typhimurium U302 strain G8430. Plasmid 2007, 57, 29–43. [Google Scholar] [CrossRef]
- Chen, C.Y.; Lindsey, R.L.; Strobaugh, T.P., Jr.; Frye, J.G.; Meinersmann, R.J. Prevalence of ColE1-like plasmids and kanamycin resistance genes in Salmonella enterica serovars. Appl. Environ. Microbiol. 2010, 76, 6707–6714. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.Y.; Strobaugh, T.P., Jr.; Frye, J.G. Characterization of small ColE1-like plasmids conferring kanamycin resistance in Salmonella enterica subsp. enterica serovars Typhimurium and Newport. Plasmid 2010, 63, 150–154. [Google Scholar] [CrossRef]
- Chen, C.Y.; Strobaugh, T.P., Jr.; Lindsey, R.L.; Frye, J.G.; Uhlich, G. Sequence analysis of a group of low molecular-weight plasmids carrying multiple IS903 elements flanking a kanamycin resistance aph gene in Salmonella enterica serovars. Plasmid 2011, 65, 246–252. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.-Y.; Strobaugh, T.P.; Nguyen, L.-H.T.; Abley, M.; Lindsey, R.L.; Jackson, C.R. Isolation and characterization of two novel groups of kanamycin-resistance ColE1-like plasmids in Salmonella enterica serotypes from food animals. PLoS ONE 2018, 13, e0193435. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McMillan, E.A.; Gupta, S.K.; Williams, L.E.; Jové, T.; Hiott, L.M.; Woodley, T.A.; Barrett, J.B.; Jackson, C.R.; Wasilenko, J.L.; Simmons, M.; et al. Antimicrobial resistance genes, cassettes, and plasmids present in Salmonella enterica associated with United States food animals. Front. Microbiol. 2019, 10, 832. [Google Scholar] [CrossRef]
- Tritt, A.; Eisen, J.A.; Facciotti, M.T.; Darling, A.E. An integrated pipeline for de novo assembly of microbial genomes. PLoS ONE 2012, 7, e42304. [Google Scholar] [CrossRef] [Green Version]
- Carattoli, A.; Zankari, E.; Garcia- Fernandez, A.; Larsen, M.V.; Lund, O.; Villa, L.; Aarestrup, F.M.; Hasman, H. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 2014, 58, 3895–3903. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Overbeek, R.; Olson, R.; Pusch, G.D.; Olsen, G.J.; Davis, J.J.; Disz, T.; Edwards, R.A.; Gerdes, S.; Parrello, B.; Shukla, M.; et al. The SEED and the rapid annotation of microbial genomes using subsystems technology (RAST). Nucleic Acids Res. 2014, 42, D206–D214. [Google Scholar] [CrossRef]
- Zankari, E.; Hasman, H.; Cosentino, S.; Vestergaard, M.; Rasmussen, S.; Lund, O.; Aarestrup, F.M.; Larsen, M.V. Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 2012, 67, 2640–2644. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Xie, Y.; Liu, M.; Tai, C.; Sun, J.; Deng, Z.; Ou, H.-Y. oriTfinder: A web-based tool for the identification of the origin of transfers in DNA sequences ofbacterial mobile genetic elements. Nucleic Acids Res. 2018, 46, W229–W234. [Google Scholar] [CrossRef] [PubMed]
- Norman, A.; Hansen, L.H.; She, Q.; Sorensen, S.J. Nucleotide sequence of pOLA52: A conjugative IncX1 plasmid from Escherichia coli which enables biofilm formation and multidrug efflux. Plasmid 2008, 60, 59–74. [Google Scholar] [CrossRef]
- Darling, A.E.; Mau, B.; Perna, N.T. progressiveMauve: Multiple genome alignment with gene gain, loss and rearrangement. PLoS ONE 2010, 5, e11147. [Google Scholar] [CrossRef] [Green Version]
- Sampei, G.; Furuya, N.; Tachibana, K.; Saitou, Y.; Suzuki, T.; Mizobuchi, K.; Komano, T. Complete genome sequence of the incompatibility group I1 plasmid R64. Plasmid 2010, 64, 92–103. [Google Scholar] [CrossRef]
- Alikhan, N.F.; Petty, N.K.; Ben Zakour, N.L.; Beatson, S.A. BLAST Ring Image Generator (BRIG): Simple prokaryote genome comparisons. BMC Genom. 2011, 12, 402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moran, R.A.; Hall, R.M. Analysis of pCERC7, a small antibiotic resistance plasmid from a commensal ST131 Escherichia coli, defines a diverse group of plasmids that include various segments adjacent to a multimer resolution site and encode the same NikA relaxase accessory protein enabling mobilisation. Plasmid 2017, 89, 42–48. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Torresen, O.K.; Star, B.; Mier, P.; Andrade-Navarro, M.A.; Bateman, A.; Jarnot, P.; Gruca, A.; Grynberg, M.; Kajava, A.V.; Promponas, V.J.; et al. Tandem repeats lead to sequence assembly errors and impose multi-level challenges for genome and protein databases. Nucleic Acids Res. 2019, 47, 10994–11006. [Google Scholar] [CrossRef] [PubMed]
| Original Isolate | Neb10β Transconjugant | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Strain | Organism | Isolation Source | Resistance Profile a | Replicon Type (PBRT) b [Sequencing] | bla Gene c | NGS d | Clone ID e | Replicon Type (PBRT) [Sequencing] | bla Gene c [Sequencing] | KanR Plasmid Mobilization f |
| 1 | S. enterica Infantis | Cattle | AMP AUG AXO CEP FOX TIO | I1 | CMY-2 | TC1d | I1 | CMY-2 | N.D. | |
| 2 | S. enterica Heidelberg | Cattle | AMP AUG AXO CEP FOX TIO | I1, X1 [ColVC] | CMY-2 | √ | LCp2 * | I1 [ColVC] | CMY-2 | All |
| 3 | S. enterica Typhimurium var. 5- | Cattle | AMP AUG AXO CEP FOX TIO | I1, FIB, X1 [FIC, ColVC] | CMY-2 | √ | TC3c | X1 | Unk [TEM-1] | pSN11/00Kan |
| 4 | S. enterica Typhimurium | Cattle | AMP AUG AXO CEP FOX TIO | I1, FIIS | CMY-2 | TC4d | I1 | CMY-2 | All | |
| 5 | S. enterica Infantis | Chicken | AMP AUG AXO FOX TIO | I1 | CMY-2 | √ | TC5f | I1 [ColVC] | CMY-2 | All |
| 6 | S. enterica Saintpaul | Turkey | AMP AUG AXO FOX TIO | I1, X1 | CMY-2 | TC6d | I1, X1 | CMY-2 | N.D. | |
| 7 | E. coli | Chicken | AMP AUG AXO COT FIS FOX GEN NAL TIO | I1, FIB, I2, K | CMY-2 | N.A. | ||||
| 8 | E. coli | Chicken | AMP AUG AXO FOX GEN TIO | FIB, B/O, K | CMY-2 | N.A. | ||||
| 10 | E. coli | Chicken | AMP AUG AXO FIS FOX GEN TIO | I1, FIB, X1, B/O, K | CMY-2 | N.A. | ||||
| 12 | E. coli | Dog | AMP AUG AXO CIP FOX GEN NAL TIO | I1 [X4, FIA, FIB, FII, Col] | CMY-2 | √ | TC12d | Unk [X4] | CMY-2 | pSN11/00Kan |
| Plasmid | Replicon Type | Size a | AR Genes | Conjugated? |
|---|---|---|---|---|
| p2I1 | I1 | 100kb | blaCMY-2 | Yes |
| p2I1-LC b | I1 | 100kb | blaCMY-2 | Yes c |
| p2X1 | X1 | 38kb | N/A | No |
| p3X1 | X1 | 43kb | blaTEM-1 | Yes |
| p3X1-TC | X1 | 43kb | blaTEM-1 | Yes (Transconjugant) |
| p3I1 | I1 | 101kb | blaCMY-2 | No |
| p5I1 | I1 | 99kb | blaCMY-2 | Yes |
| p5I1-TC | I1 | 99kb | blaCMY-2 | Yes (Transconjugant) |
| p12X4 | X4 | 37kb | blaCMY-2 | Yes |
| p12X4-TC | X4 | 37kb | blaCMY-2 | Yes (Transconjugant) |
| p12I1 | I1 | 94kb | N/A | No |
| Donor Plasmid | Mobilization Genes | Helper Strain | ||||
|---|---|---|---|---|---|---|
| LCp2 | TC3c | TC4d | TC5f | TC12d | ||
| pU302S | nikA | + | − | + | + | − |
| pSN11/00Kan | mobC-mobABD | + | + | + | + | + |
| pSe-Kan | Unknown | + | − | + | + | − |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
McMillan, E.A.; Nguyen, L.-H.T.; Hiott, L.M.; Sharma, P.; Jackson, C.R.; Frye, J.G.; Chen, C.-Y. Genomic Comparison of Conjugative Plasmids from Salmonella enterica and Escherichia coli Encoding Beta-Lactamases and Capable of Mobilizing Kanamycin Resistance Col-like Plasmids. Microorganisms 2021, 9, 2205. https://doi.org/10.3390/microorganisms9112205
McMillan EA, Nguyen L-HT, Hiott LM, Sharma P, Jackson CR, Frye JG, Chen C-Y. Genomic Comparison of Conjugative Plasmids from Salmonella enterica and Escherichia coli Encoding Beta-Lactamases and Capable of Mobilizing Kanamycin Resistance Col-like Plasmids. Microorganisms. 2021; 9(11):2205. https://doi.org/10.3390/microorganisms9112205
Chicago/Turabian StyleMcMillan, Elizabeth A., Ly-Huong T. Nguyen, Lari M. Hiott, Poonam Sharma, Charlene R. Jackson, Jonathan G. Frye, and Chin-Yi Chen. 2021. "Genomic Comparison of Conjugative Plasmids from Salmonella enterica and Escherichia coli Encoding Beta-Lactamases and Capable of Mobilizing Kanamycin Resistance Col-like Plasmids" Microorganisms 9, no. 11: 2205. https://doi.org/10.3390/microorganisms9112205
APA StyleMcMillan, E. A., Nguyen, L.-H. T., Hiott, L. M., Sharma, P., Jackson, C. R., Frye, J. G., & Chen, C.-Y. (2021). Genomic Comparison of Conjugative Plasmids from Salmonella enterica and Escherichia coli Encoding Beta-Lactamases and Capable of Mobilizing Kanamycin Resistance Col-like Plasmids. Microorganisms, 9(11), 2205. https://doi.org/10.3390/microorganisms9112205







