Campylobacter jejuni in Different Canine Populations: Characteristics and Zoonotic Potential
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Selection and Collection
2.2. Culture and Isolation of Campylobacter spp.
2.3. Isolate Identification
2.4. Antimicrobial Susceptibility Testing
2.5. Molecular Typing
2.6. Whole Genome Sequencing and Bioinformatics Analysis
2.7. Data Analysis
3. Results
3.1. Frequency and Distribution of Campylobacter spp.
3.2. Campylobacter jejuni
3.3. Comparison with Human Isolates
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- EFSA and ECDC (European Food Safety Authority and European Centre for Disease Prevention and Control). The European Union One Health 2019 Zoonoses Report. EFSA J. 2021, 19, e06406. [Google Scholar] [CrossRef]
- Mullner, P.; Spencer, S.E.F.; Wilson, D.J.; Jones, G.; Noble, A.D.; Midwinter, A.C.; Collins-Emerson, J.M.; Carter, P.; Hathaway, S.; French, N.P. Assigning the source of human campylobacteriosis in New Zealand: A comparative genetic and epidemiological approach. Infect. Genet. Evol. 2009, 9, 1311–1319. [Google Scholar] [CrossRef]
- EFSA (European Food Safety Authority). Scientific Opinion on Campylobacter in broiler meat production: Control options and performance objectives and/or targets at different stages of the food chain. EFSA J. 2011, 9, 2105. [Google Scholar] [CrossRef]
- EFSA and ECDC (European Food Safety Authority and European Centre for Disease Prevention and Control). The European Union Summary Report on Antimicrobial Resistance in zoonotic and indicator bacteria from humans, animals and food in 2018/2019. EFSA J. 2021, 19. [Google Scholar] [CrossRef]
- Acke, E. Campylobacteriosis in dogs and cats: A review. N. Z. Vet. J. 2018, 66, 221–228. [Google Scholar] [CrossRef]
- Iannino, F.; Salucci, S.; Di Donato, G.; Badagliacca, P.; Vincifori, G.; Di Giannatale, E. Campylobacter and antimicrobial resistance in dogs and humans: “One health” in practice. Vet. Ital. 2019, 55, 203–220. [Google Scholar] [CrossRef]
- Acke, E.; McGill, K.; Golden, O.; Jones, B.R.; Fanning, S.; Whyte, P. Prevalence of thermophilic Campylobacter species in household cats and dogs in Ireland. Vet. Rec. 2009, 164, 44–47. [Google Scholar] [CrossRef] [PubMed]
- Mughini-Gras, L.; Smid, J.H.; Wagenaar, J.A.; Koene, M.G.J.; Havelaar, A.H.; Friesema, I.H.M.; French, N.P.; Flemming, C.; Galson, J.D.; Graziani, C.; et al. Increased risk for Campylobacter jejuni and C. coli infection of pet origin in dog owners and evidence for genetic association between strains causing infection in humans and their pets. Epidemiol. Infect. 2013, 141, 2526–2535. [Google Scholar] [CrossRef] [Green Version]
- Rosner, B.M.; Schielke, A.; Didelot, X.; Kops, F.; Breidenbach, J.; Willrich, N.; Gölz, G.; Alter, T.; Stingl, K.; Josenhans, C.; et al. A combined case-control and molecular source attribution study of human Campylobacter infections in Germany, 2011–2014. Sci. Rep. 2017, 7, 5139. [Google Scholar] [CrossRef]
- Thépault, A.; Rose, V.; Quesne, S.; Poezevara, T.; Béven, V.; Hirchaud, E.; Touzain, F.; Lucas, P.; Méric, G.; Mageiros, L.; et al. Ruminant and chicken: Important sources of campylobacteriosis in France despite a variation of source attribution in 2009 and 2015. Sci. Rep. 2018, 8, 9305. [Google Scholar] [CrossRef]
- Kittl, S.; Heckel, G.; Korczak, B.M.; Kuhnert, P. Source attribution of human Campylobacter isolates by MLST and fla-typing and association of genotypes with quinolone resistance. PLoS ONE 2013, 8, e81796. [Google Scholar] [CrossRef] [Green Version]
- Mughini-Gras, L.; Pijnacker, R.; Coipan, C.; Mulder, A.C.; Fernandes Veludo, A.; de Rijk, S.; van Hoek, A.H.A.M.; Buij, R.; Muskens, G.; Koene, M.; et al. Sources and transmission routes of campylobacteriosis: A combined analysis of genome and exposure data. J. Infect. 2021, 82, 216–226. [Google Scholar] [CrossRef]
- Wolfs, T.F.W.; Duim, B.; Geelen, S.P.M.; Rigter, A.; Thomson-carter, F.; Wagenaar, J.A. Neonatal Sepsis by Campylobacter jejuni: Genetically Proven Transmission from a Household Puppy. Clin. Infect. Dis. 2001, 32, 97–99. [Google Scholar] [CrossRef]
- Damborg, P.; Olsen, K.E.P.; Nielsen, E.M.; Guardabassi, L. Occurrence of Campylobacter jejuni in Pets Living with Human Patients Infected with C. jejuni. J. Clin. Microbiol. 2004, 42, 1363–1364. [Google Scholar] [CrossRef] [Green Version]
- McEwen, S.A.; Collignon, P.J. Antimicrobial Resistance: A One Health Perspective. Antimicrob. Resist. Bact. Livest. Companion Anim. 2018, 6, 521–547. [Google Scholar] [CrossRef] [Green Version]
- Yamazaki-Matsune, W.; Taguchi, M.; Seto, K.; Kawahara, R.; Kawatsu, K.; Kumeda, Y.; Kitazato, M.; Nukina, M.; Misawa, N.; Tsukamoto, T. Development of a multiplex PCR assay for identification of Campylobacter coli, Campylobacter fetus, Campylobacter hyointestinalis subsp. hyointestinalis, Campylobacter jejuni, Campylobacter lari and Campylobacter upsaliensis. J. Med. Microbiol. 2007, 56, 1467–1473. [Google Scholar] [CrossRef] [Green Version]
- The European Committee on Antimicrobial Susceptibility Testing. Breakpoint Tables for Interpretation of MICs and Zone Diameters. Version 11.0. 2021. Available online: http://www.eucast.org (accessed on 1 September 2021).
- Versalovic, J.; Koeuth, T.; Lupski, J.R.; Plaza, O.B. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. 1991, 19, 6823–6831. [Google Scholar] [CrossRef]
- Silveira, L.; Nunes, A.; Pista, A.; Isidro, J.; Belo Correia, C.; Saraiva, M.; Batista, R.; Castanheira, I.; MacHado, J.; Gomes, J.P. Characterization of Multidrug-Resistant Isolates of Salmonella enterica Serovars Heidelberg and Minnesota from Fresh Poultry Meat Imported to Portugal. Microb. Drug Resist. 2021, 27, 87–98. [Google Scholar] [CrossRef]
- Llarena, A.; Ribeiro-Gonçalves, B.F.; Nuno Silva, D.; Halkilahti, J.; Machado, M.P.; Da Silva, M.S.; Jaakkonen, A.; Isidro, J.; Hämäläinen, C.; Joenperä, J.; et al. INNUENDO: A cross-sectoral platform for the integration of genomics in the surveillance of food-borne pathogens. EFSA Support. Publ. 2018, 15, 1498E. [Google Scholar] [CrossRef]
- Aziz, R.K.; Bartels, D.; Best, A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST Server: Rapid annotations using subsystems technology. BMC Genom. 2008, 9, 75. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rossi, M.; da Silva, M.S.; Ribeiro-Gonçalves, B.F.; Silva, D.N.; Machado, M.P.; Oleastro, M.; Borges, V.; Isidro, J.; Viera, L.; Barker, D.O.; et al. INNUENDO whole genome and core genome MLST schemas and datasets for Campylobacter jejuni. Zenodo 2018, 4. [Google Scholar] [CrossRef]
- Silva, M.; Machado, M.P.; Silva, D.N.; Rossi, M.; Moran-Gilad, J.; Santos, S.; Ramirez, M.; Carriço, J.A. chewBBACA: A complete suite for gene-by-gene schema creation and strain identification. Microb. Genom. 2018, 4, e000166. [Google Scholar] [CrossRef]
- Francisco, A.P.; Bugalho, M.; Ramirez, M.; Carriço, J.A. Global optimal eBURST analysis of multilocus typing data using a graphic matroid approach. BMC Bioinform. 2009, 10, 152. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ribeiro-Gonçalves, B.; Francisco, A.P.; Vaz, C.; Ramirez, M.; Carriço, J.A. PHYLOViZ Online: Web-based tool for visualization, phylogenetic inference, analysis and sharing of minimum spanning trees. Nucleic Acids Res. 2016, 44, 246–251. [Google Scholar] [CrossRef]
- Giacomelli, M.; Follador, N.; Coppola, L.M.; Martini, M.; Piccirillo, A. Survey of Campylobacter spp. in owned and unowned dogs and cats in Northern Italy. Vet. J. 2015, 204, 333–337. [Google Scholar] [CrossRef]
- Amar, C.; Kittl, S.; Spreng, D.; Thomann, A.; Korczak, B.M.; Burnens, A.P.; Kuhnert, P. Genotypes and antibiotic resistance of canine Campylobacter jejuni isolates. Vet. Microbiol. 2014, 168, 124–130. [Google Scholar] [CrossRef]
- Chaban, B.; Ngeleka, M.; Hill, J.E. Detection and quantification of 14 Campylobacter species in pet dogs reveals an increase in species richness in feces of diarrheic animals. BMC Microbiol. 2010, 10, 73. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carbonero, A.; Torralbo, A.; Borge, C.; García-Bocanegra, I.; Arenas, A.; Perea, A. Campylobacter spp., C. Jejuni and C. Upsaliensis infection-associated factors in healthy and ill dogs from clinics in Cordoba, Spain. Screening tests for antimicrobial susceptibility. Comp. Immunol. Microbiol. Infect. Dis. 2012, 35, 505–512. [Google Scholar] [CrossRef]
- Man, S.M. The clinical importance of emerging Campylobacter species. Nat. Rev. Gastroenterol. Hepatol. 2011, 8, 669–685. [Google Scholar] [CrossRef]
- Bourke, B.; Chan, V.L.; Sherman, P. Campylobacter upsaliensis: Waiting in the wings. Clin. Microbiol. Rev. 1998, 11, 440–449. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aspinall, S.T.; Wareing, D.R.A.; Hayward, P.G.; Hutchinson, D.N. Selective medium for thermophilic campylobacters including Campylobacter upsaliensis. J. Clin. Pathol. 1993, 46, 829–831. [Google Scholar] [CrossRef] [Green Version]
- Bojanić, K.; Midwinter, A.C.; Marshall, J.C.; Biggs, P.J.; Acke, E. Isolation of emerging Campylobacter species in working farm dogs and their frozen home-killed raw meat diets. J. Vet. Diagn. Investig. 2019, 31, 23–32. [Google Scholar] [CrossRef] [Green Version]
- Aspinall, S.T.; Wareing, D.R.A.; Hayward, P.G.; Hutchinson, D.N. A comparison of a new campylobacter selective medium (CAT) with membrane filtration for the isolation of thermophilic campylobacters including Campylobacter upsaliensis. J. Appl. Bacteriol. 1996, 80, 645–650. [Google Scholar] [CrossRef]
- Santaniello, A.; Varriale, L.; Dipineto, L.; Borrelli, L.; Pace, A.; Fioretti, A.; Menna, L.F. Presence of campylobacter jejuni and C. Coli in dogs under training for animal-assisted therapies. Int. J. Environ. Res. Public Health 2021, 18, 3717. [Google Scholar] [CrossRef]
- Lee, M.K.; Billington, S.J.; Joens, L.A. Potential virulence and antimicrobial susceptibility of Campylobacter jejuni isolates from food and companion animals. Foodborne Pathog. Dis. 2004, 1, 223–230. [Google Scholar] [CrossRef]
- Rossi, M.; Hänninen, M.L.; Revez, J.; Hannula, M.; Zanoni, R.G. Occurrence and species level diagnostics of Campylobacter spp., enteric Helicobacter spp. and Anaerobiospirillum spp. in healthy and diarrheic dogs and cats. Vet. Microbiol. 2008, 129, 304–314. [Google Scholar] [CrossRef]
- Sahin, O.; Burrough, E.R.; Pavlovic, N.; Frana, T.S.; Madson, D.M.; Zhang, Q. Campylobacter jejuni as a cause of canine abortions in the United States. J. Vet. Diagn. Investig. 2014, 26, 699–704. [Google Scholar] [CrossRef] [Green Version]
- Revez, J.; Rossi, M.; Ellström, P.; de Haan, C.; Rautelin, H.; Hänninen, M.L. Finnish Campylobacter jejuni strains of multilocus sequence type ST-22 complex have two lineages with different characteristics. PLoS ONE 2011, 6, e26880. [Google Scholar] [CrossRef]
- Islam, Z.; van Belkum, A.; Wagenaar, J.A.; Cody, A.J.; de Boer, A.G.; Tabor, H.; Jacobs, B.C.; Talukder, K.A.; Endtz, H.P. Comparative genotyping of Campylobacter jejuni strains from patients with Guillain-Barré syndrome. PLoS ONE 2009, 4, e7257. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taboada, E.N.; van Belkum, A.; Yuki, N.; Acedillo, R.R.; Godschalk, P.C.R.; Koga, M.; Endtz, H.P.; Gilbert, M.; Nash, J.H.E. Comparative genomic analysis of Campylobacter jejuni associated with Guillain-Barré and Miller Fisher syndromes: Neuropathogenic and enteritis-associated isolates can share high levels of genomic similarity. BMC Genom. 2007, 8, 359. [Google Scholar] [CrossRef] [Green Version]
- Dingle, K.E.; Colles, F.M.; Ure, R.; Wagenaar, J.A.; Duim, B.; Bolton, F.J.; Fox, A.J.; Wareing, D.R.A.; Maiden, M.C.J. Characterization of Campylobacter jejuni Clones: A Basis for Epidemiologic Investigation. Emerg. Infect. Dis. 2002, 8, 949–955. [Google Scholar] [CrossRef] [PubMed]
- De Haan, C.P.A.; Kivistö, R.; Hakkinen, M.; Rautelin, H.; Hänninen, M.L. Decreasing trend of overlapping multilocus sequence types between human and chicken campylobacter jejuni isolates over a decade in finland. Appl. Environ. Microbiol. 2010, 76, 5228–5236. [Google Scholar] [CrossRef] [PubMed] [Green Version]



| N-Total [n1-Companion/n2-Hunting] | Gender | Age | Diarrhea | Setting | ||||
|---|---|---|---|---|---|---|---|---|
| Male | Female | 0–2 | >2 | Yes | No | Urban | Rural | |
| N. dogs | 55 | 55 | 42 | 68 | 12 | 98 | 50 | 60 |
| [32/23] | [39/16] | [23/19] | [44/24] | [12/0] | [59/39] | [50/0] | [21/39] | |
| N. samples | 63 | 62 | 47 | 78 | 12 | 113 | 50 | 75 |
| [32/31] | [39/23] | [23/24] | [44/34] | [12/0] | [59/54] | [50/0] | [21/54] | |
| Distribution by species | ||||||||
| C. jejuni | 10 | 4 | 4 | 10 | 1 | 13 | 1 | 13 |
| [2/8] | [0/4] | [2/2] | [0/10] | [1/0] | [1/12] | [1/0] | [1/12] | |
| C. upsaliensis | 2 | 2 | 2 | 2 | 3 | 1 | 3 | 1 |
| [2/0] | [2/0] | [2/0] | [2/0] | [3/0] | [1/0] | [3/0] | [1/0] | |
| C. lari | 6 | 7 | 5 | 8 | 0 | 13 | 0 | 13 |
| [0/6] | [0/7] | [0/5] | [0/8] | [0/0] | [0/13] | [0/0] | [0/13] | |
| C. coli | 1 | 0 | 1 | 0 | 0 | 1 | 0 | 1 |
| [1/0] | [0/0] | [1/0] | [0/0] | [0/0] | [1/0] | [0/0] | [1/0] | |
| Total | 19 | 13 | 12 | 20 | 4 | 28 | 4 | 28 |
| [5/14] | [2/11] | [5/7] | [2/18] | [4/0] | [3/25] | [4/0] | [3/25] | |
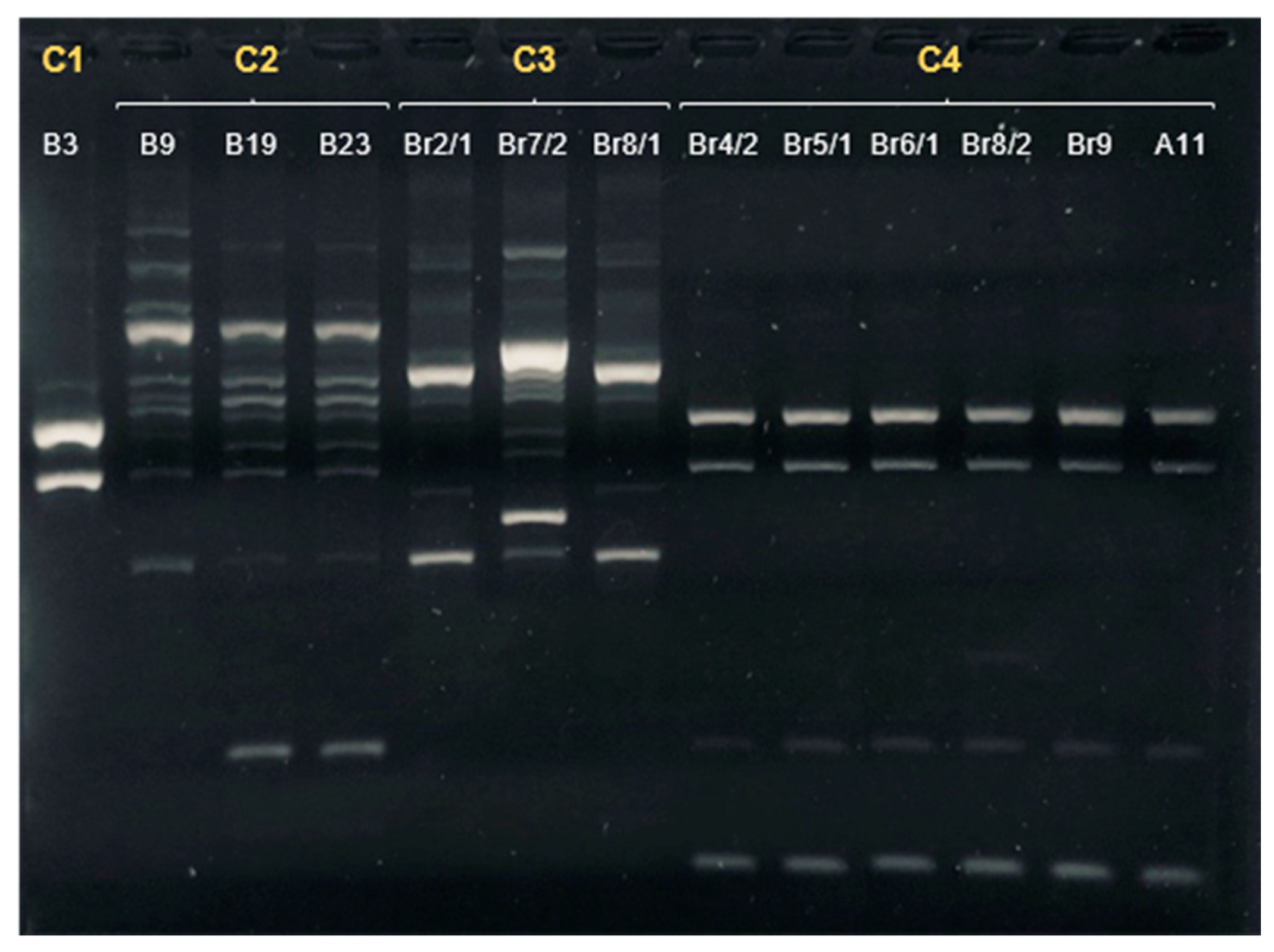
| Sample | Date of Isolation | Group | ERIC-PCR Cluster | MLST Sequence Type | Clonal Complex | flaA svr |
|---|---|---|---|---|---|---|
| A11 | 24/09/2020 | Companion/Urban | 4 | 22 | 22 | 161 |
| B3 | 23/10/2020 | Hunting/Rural | 1 | 48 | 48 | 32;102 |
| B9 | 23/10/2020 | Hunting/Rural | 2 | 8569 | 179 | 571 |
| B19 | 6/11/2020 | Hunting/Rural | 2 | 148 | 21 | 36;36 |
| Br2/1 | 20/11/2020 | Hunting/Rural | 3 | 6461 | 353 | 67 |
| Br4/2 | 20/11/2020 | Hunting/Rural | 4 | 22 | 22 | 161 |
| Br7/2 | 20/11/2020 | Hunting/Rural | 3 | 6461 | 353 | 49;395 |
| Br8/1 | 20/11/2020 | Hunting/Rural | 3 | 6461 | 353 | 49 |
| Br8/2 | 20/11/2020 | Hunting/Rural | 4 | 22 | 22 | 161 |
| Br9 | 20/11/2020 | Hunting/Rural | 4 | 22 | 22 | 161 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lemos, M.-L.; Nunes, A.; Ancora, M.; Cammà, C.; Costa, P.M.d.; Oleastro, M. Campylobacter jejuni in Different Canine Populations: Characteristics and Zoonotic Potential. Microorganisms 2021, 9, 2231. https://doi.org/10.3390/microorganisms9112231
Lemos M-L, Nunes A, Ancora M, Cammà C, Costa PMd, Oleastro M. Campylobacter jejuni in Different Canine Populations: Characteristics and Zoonotic Potential. Microorganisms. 2021; 9(11):2231. https://doi.org/10.3390/microorganisms9112231
Chicago/Turabian StyleLemos, Maria-Leonor, Alexandra Nunes, Massimo Ancora, Cesare Cammà, Paulo Martins da Costa, and Mónica Oleastro. 2021. "Campylobacter jejuni in Different Canine Populations: Characteristics and Zoonotic Potential" Microorganisms 9, no. 11: 2231. https://doi.org/10.3390/microorganisms9112231
APA StyleLemos, M.-L., Nunes, A., Ancora, M., Cammà, C., Costa, P. M. d., & Oleastro, M. (2021). Campylobacter jejuni in Different Canine Populations: Characteristics and Zoonotic Potential. Microorganisms, 9(11), 2231. https://doi.org/10.3390/microorganisms9112231







