Dynamic Structure of Yeast Septin by Fast Fluctuation-Enhanced Structured Illumination Microscopy
Abstract
:1. Introduction
2. Materials and Methods
2.1. Yeast Strains
2.2. Plasmid for Mammalian Cells
2.3. Imaging Dish Preparation
2.4. Yeast Cell Preparation
2.5. Mammalian Cell Culture
2.6. Imaging Acquisition and Analysis
3. Results
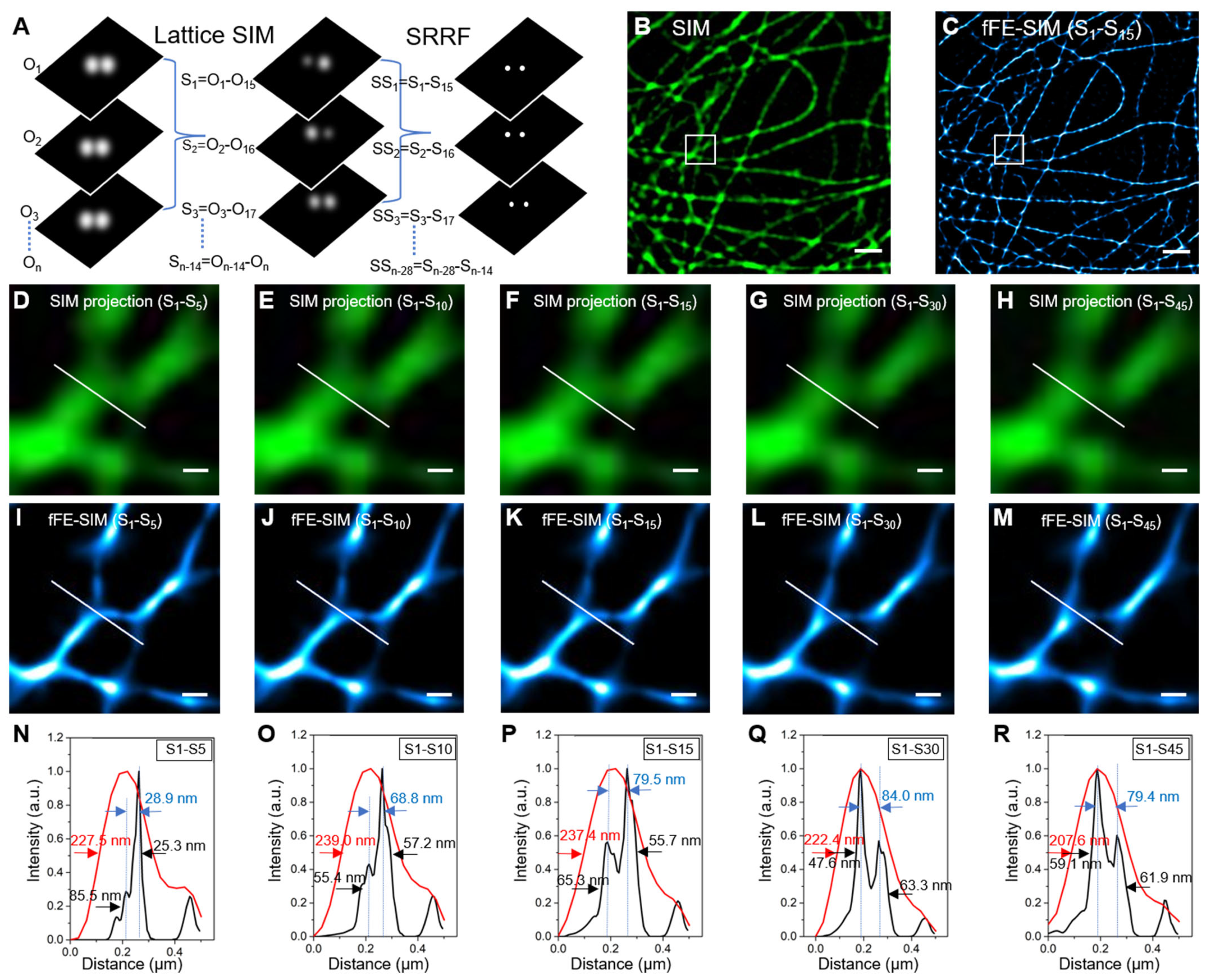
3.1. Lateral Resolution Verification of fFE-SIM
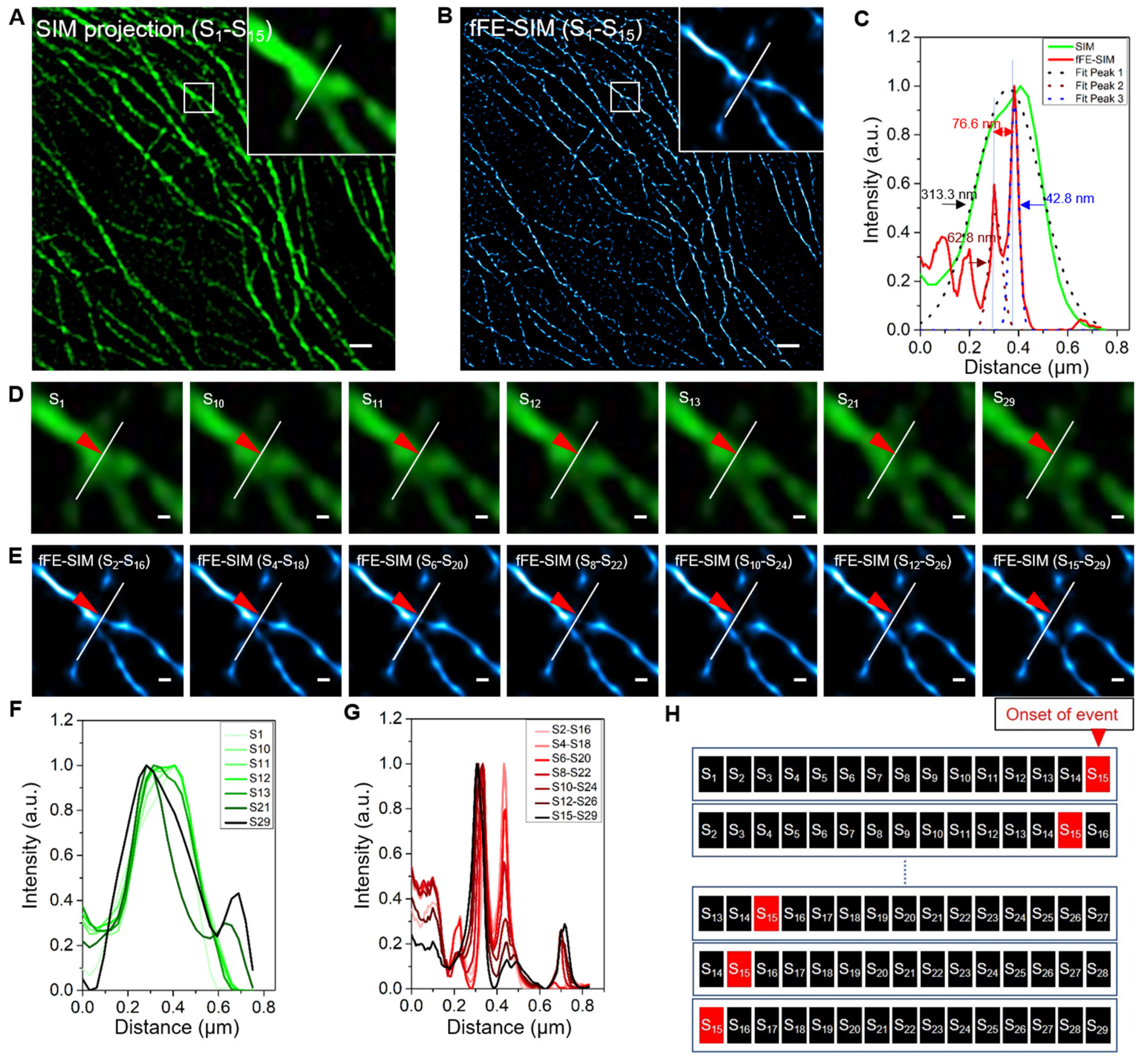
3.2. Lateral Resolution Verification of fFE-SIM in Live Cell
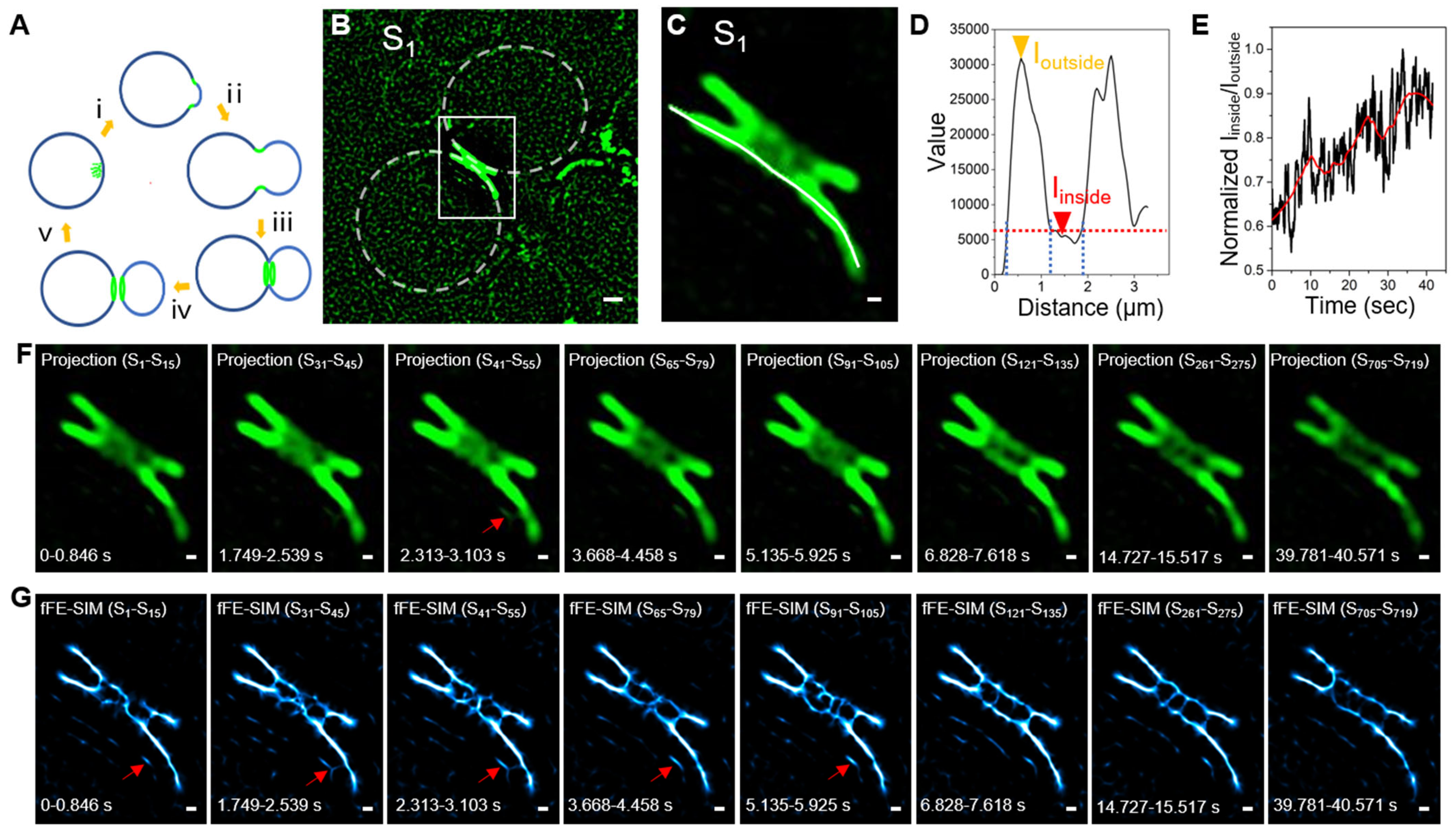
3.3. Long Term Living Yeast Cell Imaging of a Cdc12 Labeled Septin Ring by fFE-SIM
3.4. A New Architectural Model of a Cdc12 Labeled Septin Ring
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Park, H.-O.; Bi, E. Central Roles of Small GTPases in the Development of Cell Polarity in Yeast and Beyond. Microbiol. Mol. Biol. Rev. 2007, 71, 48–96. [Google Scholar] [CrossRef] [Green Version]
- Hartwell, L. Genetic control of the cell division cycle in yeast *1IV. Genes controlling bud emergence and cytokinesis. Exp. Cell Res. 1971, 69, 265–276. [Google Scholar] [CrossRef]
- Versele, M.; Thorner, J. Some assembly required: Yeast septins provide the instruction manual. Trends Cell Biol. 2005, 15, 414–424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fares, H.; Goetsch, L.; Pringle, J.R. Identification of a developmentally regulated septin and involvement of the septins in spore formation in Saccharomyces cerevisiae. J. Cell Biol. 1996, 132, 399–411. [Google Scholar] [CrossRef] [Green Version]
- De Virgilio, C.; DeMarini, D.J.; Pringle, J.R. SPR28, a sixth member of the septin gene family in Saccharomyces cerevisiae that is expressed specifically in sporulating cells. Microbiology 1996, 142, 2897–2905. [Google Scholar] [CrossRef] [Green Version]
- Szkotnicki, L.; Crutchley, J.M.; Zyla, T.R.; Bardes, E.S.; Lew, D.J. The Checkpoint Kinase Hsl1p Is Activated by Elm1p-dependent Phosphorylation. Mol. Biol. Cell 2008, 19, 4675–4686. [Google Scholar] [CrossRef] [Green Version]
- Berlin, A.; Paoletti, A.; Chang, F. Mid2p stabilizes septin rings during cytokinesis in fission yeast. J. Cell Biol. 2003, 160, 1083–1092. [Google Scholar] [CrossRef] [Green Version]
- Versele, M.; Gullbrand, B.; Shulewitz, M.J.; Cid, V.J.; Bahmanyar, S.; Chen, R.E.; Barth, P.; Alber, T.; Thorner, J. Protein–Protein Interactions Governing Septin Heteropentamer Assembly and Septin Filament Organization in Saccharomyces cerevisiae. Mol. Biol. Cell 2004, 15, 4568–4583. [Google Scholar] [CrossRef] [Green Version]
- Ong, K.; Svitkina, T.; Bi, E. Visualization of in vivo septin ultrastructures by platinum replica electron microscopy. Methods Cell Biol. 2016, 136, 73–97. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Byers, B.; Goetsch, L. A highly ordered ring of membrane-associated filaments in budding yeast. J. Cell Biol. 1976, 69, 717–721. [Google Scholar] [CrossRef] [PubMed]
- Hayat, M.E. Fixation for Electron Microscopy; Elsevier: Amsterdam, The Netherlands, 2012. [Google Scholar]
- Miller, S.E.; Goldsmith, C.S. Caution in Identifying Coronaviruses by Electron Microscopy. J. Am. Soc. Nephrol. 2020, 31, 2223–2224. [Google Scholar] [CrossRef]
- Vrabioiu, A.M.; Mitchison, T.J. Structural insights into yeast septin organization from polarized fluorescence microscopy. Nat. Cell Biol. 2006, 443, 466–469. [Google Scholar] [CrossRef]
- DeMay, B.S.; Bai, X.; Howard, L.; Occhipinti, P.; Meseroll, R.; Spiliotis, E.T.; Oldenbourg, R.; Gladfelter, A.S. Septin filaments exhibit a dynamic, paired organization that is conserved from yeast to mammals. J. Cell Biol. 2011, 193, 1065–1081. [Google Scholar] [CrossRef] [Green Version]
- Zhanghao, K.; Chen, L.; Yang, X.-S.; Wang, M.-Y.; Jing, Z.-L.; Han, H.-B.; Zhang, M.Q.; Jin, D.; Gao, J.-T.; Xi, P. Super-resolution dipole orientation mapping via polarization demodulation. Light Sci. Appl. 2016, 5, e16166. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Wang, M.; Zhang, X.; Zhang, M.; Hu, Y.; Shi, Z.; Xi, P.; Gao, J. Group-sparsity-based super-resolution dipole orientation mapping. IEEE Trans. Med. Imaging 2019, 38, 2687–2694. [Google Scholar] [CrossRef]
- Ong, K.; Wloka, C.; Okada, S.; Svitkina, T.; Bi, E. Architecture and dynamic remodelling of the septin cytoskeleton during the cell cycle. Nat. Commun. 2014, 5, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caviston, J.P.; Longtine, M.; Pringle, J.R.; Bi, E. The Role of Cdc42p GTPase-activating Proteins in Assembly of the Septin Ring in Yeast. Mol. Biol. Cell 2003, 14, 4051–4066. [Google Scholar] [CrossRef] [Green Version]
- Hu, Q.; Nelson, W.J.; Spiliotis, E.T. Forchlorfenuron Alters Mammalian Septin Assembly, Organization and Dynamics. J. Biol. Chem. 2008, 283, 29563–29571. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rust, M.J.; Bates, M.; Zhuang, X. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM). Nat. Methods 2006, 3, 793–796. [Google Scholar] [CrossRef] [Green Version]
- Klar, T.; Hell, S.W. Subdiffraction resolution in far-field fluorescence microscopy. Opt. Lett. 1999, 24, 954–956. [Google Scholar] [CrossRef] [PubMed]
- Betzig, E.; Patterson, G.H.; Sougrat, R.; Lindwasser, O.W.; Olenych, S.; Bonifacino, J.S.; Davidson, M.W.; Lippincott-Schwartz, J.; Hess, H.F. Imaging Intracellular Fluorescent Proteins at Nanometer Resolution. Science 2006, 313, 1642–1645. [Google Scholar] [CrossRef] [Green Version]
- Huang, X.; Fan, J.; Li, L.; Liu, H.; Wu, R.; Wu, Y.; Wei, L.; Mao, H.; Lal, A.; Xi, P.; et al. Fast, long-term, super-resolution imaging with Hessian structured illumination microscopy. Nat. Biotechnol. 2018, 36, 451–459. [Google Scholar] [CrossRef]
- Betzig, E. Excitation strategies for optical lattice microscopy. Opt. Express 2005, 13, 3021–3036. [Google Scholar] [CrossRef]
- Gustafsson, M.G.L. Surpassing the lateral resolution limit by a factor of two using structured illumination microscopy. SHORT COMMUNICATION. J. Microsc. 2000, 198, 82–87. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, B.; Yao, L.; Jing, Y.; Fei, Y.; Bai, Q.; Mi, L.; Ma, J. Multicomposite super-resolution microscopy: Enhanced Airyscan resolution with radial fluctuation and sample expansions. J. Biophotonics 2020, 13, e2419. [Google Scholar] [CrossRef] [PubMed]
- Gustafsson, N.; Culley, S.; Ashdown, G.; Owen, G.A.D.M.; Pereira, P.M.; Henriques, N.G.S.C.P.M.P.R. Fast live-cell conventional fluorophore nanoscopy with ImageJ through super-resolution radial fluctuations. Nat. Commun. 2016, 7, 12471. [Google Scholar] [CrossRef] [Green Version]
- Han, Y.; Lu, X.; Zhang, Z.; Liu, W.; Chen, Y.; Liu, X.; Hao, X.; Kuang, C. Ultra-fast, universal super-resolution radial fluctuations (SRRF) algorithm for live-cell super-resolution microscopy. Opt Express. 2019, 27, 38337–38348. [Google Scholar] [CrossRef]
- Saarikangas, J.; Barral, Y. The emerging functions of septins in metazoans. EMBO Rep. 2011, 12, 1118–1126. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roeseler, S.; Sandrock, K.; Bartsch, I.; Zieger, B. Septins, a Novel Group of GTP-binding Proteins—Relevance in Hemostasis, Neuropathology and Oncogenesis. Klinische Pädiatr. 2009, 221, 150–155. [Google Scholar] [CrossRef]
- Cannon, K.S.; Woods, B.L.; Crutchley, J.M.; Gladfelter, A.S. An amphipathic helix enables septins to sense micrometer-scale membrane curvature. J. Cell Biol. 2019, 218, 1128–1137. [Google Scholar] [CrossRef] [Green Version]
- Woods, B.L.; Gladfelter, A.S. The state of the septin cytoskeleton from assembly to function. Curr. Opin. Cell Biol. 2021, 68, 105–112. [Google Scholar] [CrossRef] [PubMed]
- Palmer, R.E.; Sullivan, D.S.; Huffaker, T.; Koshland, D. Role of astral microtubules and actin in spindle orientation and migration in the budding yeast, Saccharomyces cerevisiae. J. Cell Biol. 1992, 119, 583–593. [Google Scholar] [CrossRef] [PubMed] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yao, L.; Zhang, L.; Chen, L.; Gong, X.; Zhong, J.; Wang, B.; Fei, Y.; Mi, L.; Ma, J. Dynamic Structure of Yeast Septin by Fast Fluctuation-Enhanced Structured Illumination Microscopy. Microorganisms 2021, 9, 2255. https://doi.org/10.3390/microorganisms9112255
Yao L, Zhang L, Chen L, Gong X, Zhong J, Wang B, Fei Y, Mi L, Ma J. Dynamic Structure of Yeast Septin by Fast Fluctuation-Enhanced Structured Illumination Microscopy. Microorganisms. 2021; 9(11):2255. https://doi.org/10.3390/microorganisms9112255
Chicago/Turabian StyleYao, Longfang, Li Zhang, Liwen Chen, Xingyu Gong, Jiahui Zhong, Baoju Wang, Yiyan Fei, Lan Mi, and Jiong Ma. 2021. "Dynamic Structure of Yeast Septin by Fast Fluctuation-Enhanced Structured Illumination Microscopy" Microorganisms 9, no. 11: 2255. https://doi.org/10.3390/microorganisms9112255
APA StyleYao, L., Zhang, L., Chen, L., Gong, X., Zhong, J., Wang, B., Fei, Y., Mi, L., & Ma, J. (2021). Dynamic Structure of Yeast Septin by Fast Fluctuation-Enhanced Structured Illumination Microscopy. Microorganisms, 9(11), 2255. https://doi.org/10.3390/microorganisms9112255





