Intestinal Dominance by Serratia marcescens and Serratia ureilytica among Neonates in the Setting of an Outbreak
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection from Neonates during the Thirteen Weeks of the Outbreak
2.2. Water Sample Collection
2.3. DNA Extraction
2.4. Relative Quantification of the Intestinal Loads of Serratia Species
2.5. Clonality Analysis
2.6. Whole-Genome Sequencing
2.7. Retrieving Clinical Microbiology Records
3. Results
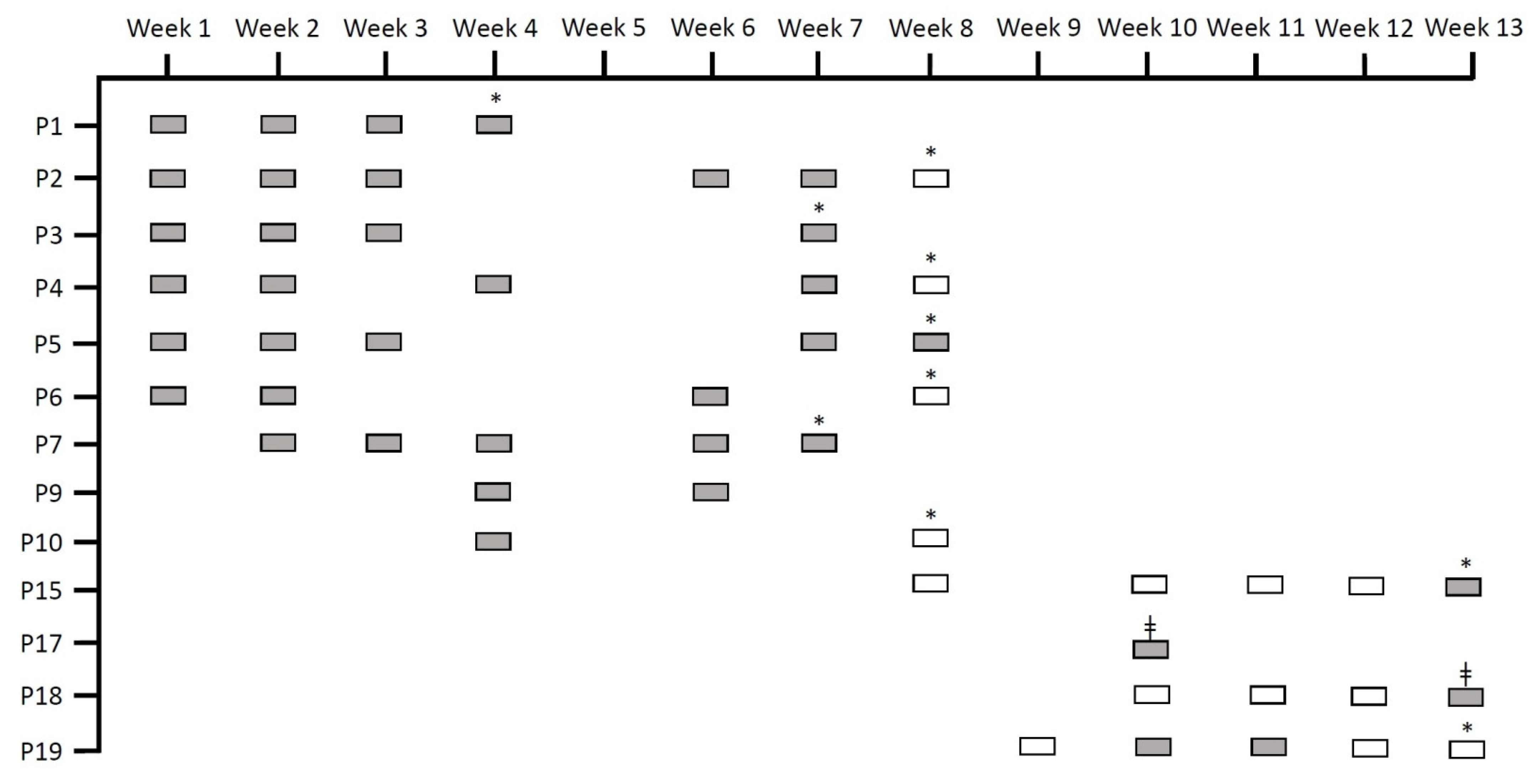
3.1. Patients, Outbreaks, and Samples
3.2. Random Amplified Polymorphic DNA (RAPD) Analysis
3.3. Whole-Genome Sequencing
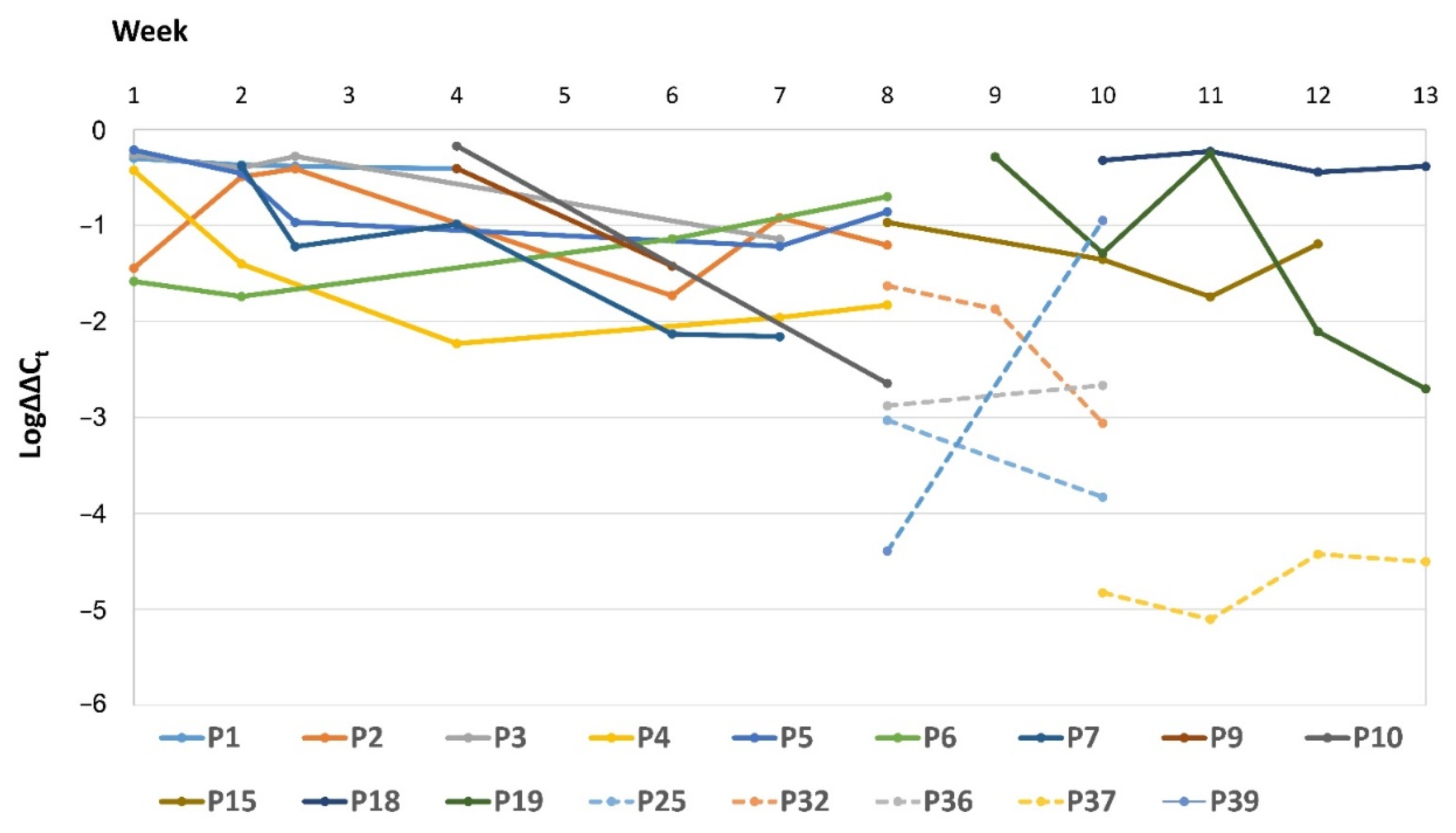
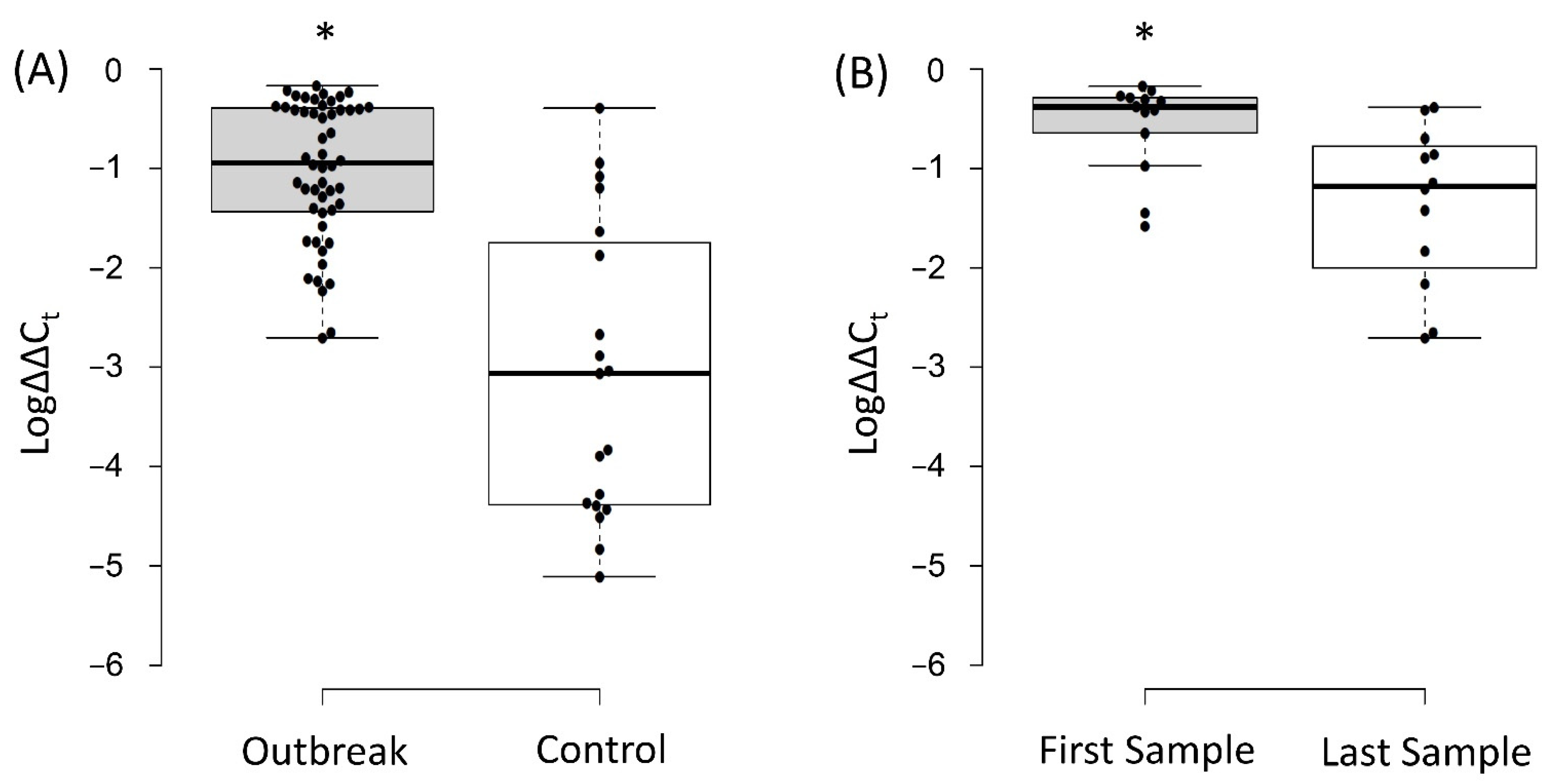
3.4. Relative Quantification of the Intestinal Load of Serratia spp.
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Mahlen, S.D. Serratia infections: From military experiments to current practice. Clin. Microbiol. Rev. 2011, 24, 755–791. [Google Scholar] [CrossRef]
- Quinn, L.; Ailsworth, M.; Matthews, E.; Kellams, A.; Shirley, D.A. Serratia marcescens Colonization Causing Pink Breast Milk and Pink Diapers: A Case Report and Literature Review. Breastfeed. Med. 2018, 13, 388–394. [Google Scholar] [CrossRef] [PubMed]
- Ottolini, K.M.; Litke-Wager, C.A.; Johnson, R.L.; Schulz, E.V. Serratia Chorioamnionitis and Culture Proven Sepsis in a Preterm Neonate: A Case Report and Review of the Literature. Pediatric Infect. Dis. J. 2020, 40, e62–e65. [Google Scholar] [CrossRef] [PubMed]
- The European Committee on Antimicrobial Susceptibility Testing (EUCAST). Expert Rules on Enterobacterales. Version 3.2. 2021. Available online: https://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Expert_Rules/2021/ExpertRules_V3.2_20211018_EnterobactEntero.pdf (accessed on 19 October 2021).
- Fisher, M.; Diago-Navarro, E.; Kaplun, O.; Sin, E.; Fries, B.C. Serratia marcescens Strains Carrying blaKPC-2 and blaKPC-3 Carbapenemase Associated With Chronic Mechanical Ventilation. Open Forum Infect. Dis. 2018, 5, S362. [Google Scholar] [CrossRef][Green Version]
- Rohit, A.; Suresh Kumar, D.; Dhinakaran, I.; Joy, J.; Kumar, D.; Ballamoole, K.K.; Karunasagar, I.; Karola, P.; Dag, H. Whole-genome-based analysis reveals multiclone Serratia marcescens outbreaks in a non-Neonatal Intensive Care Unit setting in a tertiary care hospital in India. J. Med. Microbiol. 2019, 68, 616–621. [Google Scholar] [CrossRef] [PubMed]
- Viau, R.; Frank, K.M.; Jacobs, M.R.; Wilson, B.; Kaye, K.; Donskey, C.J.; Perez, F.; Endimiani, A.; Bonomo, R.A. Intestinal Carriage of Carbapenemase-Producing Organisms: Current Status of Surveillance Methods. Clin. Microbiol. Rev. 2016, 29, 1–27. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Lin, S.; Kelen, G.D.; Quinn, T.C.; Dick, J.D.; Gaydos, C.A.; Rothman, R.E. Quantitative multiprobe PCR assay for the simultaneous detection and species identification of bacterial pathogens. J. Clin. Microbiol. 2002, 40, 3449–3454. [Google Scholar] [CrossRef] [PubMed]
- Joyner, J.; Wanless, D.; Sinigalliano, C.D.; Lipp, E.K. Use of Quantitative Real-Time PCR for Direct Detection of Serratia marcescens in Marine and Other Aquatic Environments. Appl. Environ. Microbiol. 2014, 80, 1679–1683. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Svec, D.; Tichopad, A.; Novosadova, V.; Pfaffl, M.W.; Kubista, M. How good is a PCR efficiency estimate: Recommendations for precise and robust qPCR efficiency assessments. Biomol. Detect. Quantif. 2015, 3, 9–16. [Google Scholar] [CrossRef]
- Hsueh, P.R.; Teng, L.J.; Yang, P.C.; Chen, Y.C.; Ho, S.W.; Luh, K.T. Persistence of a multidrug-resistant Pseudomonas aeruginosa clone in an intensive care burn unit. J. Clin. Microbiol. 1998, 36, 1347–1351. [Google Scholar] [CrossRef] [PubMed]
- Seemann, T.; Goncalves da Silva, A.; Bulach, D.M.; Schultz, M.B.; Kwong, J.C.; Howden, B.P. Nullarbor Github. Available online: https://github.com/tseemann/nullarbor (accessed on 19 July 2021).
- The European Committee on Antimicrobial Susceptibility Testing (EUCAST). Breakpoint Tables for Interpretation of MICs and Zone Diameters. Version 9.0. 2019. Available online: http://www.eucast.org (accessed on 15 June 2021).
- Petersen, L.M.; Tisa, L.S. Friend or foe? A review of the mechanisms that drive Serratia towards diverse lifestyles. Can. J. Microbiol. 2013, 59, 627–640. [Google Scholar] [CrossRef] [PubMed]
- Firmo, E.F.; Beltrão, E.M.B.; Silva, F.R.F.D.; Alves, L.C.; Brayner, F.A.; Veras, D.L.; Lopes, A.C.S. Association of blaNDM-1 with blaKPC-2 and aminoglycoside-modifying enzyme genes among Klebsiella pneumoniae, Proteus mirabilis and Serratia marcescens clinical isolates in Brazil. J. Glob. Antimicrob. Resist. 2020, 21, 255–261. [Google Scholar] [CrossRef] [PubMed]
- Bhadra, B.; Roy, P.; Chakraborty, R. Serratia ureilytica sp. nov., a novel urea-utilizing species. Int. J. Syst. Evol. Microbiol. 2005, 55, 2155–2158. [Google Scholar] [CrossRef]
- Bourdin, T.; Monnier, A.; Benoit, M.È.; Bédard, E.; Prévost, M.; Quach, C.; Déziel, E.; Constant, P. A High-Throughput Short Sequence Typing Scheme for Serratia marcescens Pure Culture and Environmental DNA. Appl. Environ. Microbiol. 2021, AEM0139921. [Google Scholar] [CrossRef]
- Young, V.B. The role of the microbiome in human health and disease: An introduction for clinicians. BMJ 2017, 356, j831. [Google Scholar] [CrossRef] [PubMed]
- Van Loon, K.; Voor in’t Holt, A.F.; Vos, M.C. A Systematic Review and Meta-analyses of the Clinical Epidemiology of Carbapenem-Resistant Enterobacteriaceae. Antimicrob. Agents Chemother. 2017, 62, e01730-17. [Google Scholar] [CrossRef] [PubMed]
- Weinstein, R.A. Intensive care unit environments and the fecal patina: A simple problem? Crit. Care Med. 2012, 40, 1333–1334. [Google Scholar] [CrossRef]
- Escribano, E.; Saralegui, C.; Moles, L.; Montes, M.T.; Alba, C.; Alarcón, T.; Lázaro-Perona, F.; Rodríguez, J.M.; Sáenz de Pipaón, M.; Del Campo, R. Influence of a Serratia marcescens outbreak on the gut microbiota establishment process in low-weight preterm neonates. PLoS ONE 2019, 21, e0216581. [Google Scholar] [CrossRef] [PubMed]


| Group 1 Neonates | Group 2 Neonates | |
|---|---|---|
| Number of Patients | 13 | 29 |
| Days of Life Upon Inclusion (Mean ± SD) | 17.5 ± 7.1 | 23.5 ± 36.9 |
| GA, weeks (Mean ± SD) | 32.2 ± 4.9 | 33.2 ± 5.1 |
| Average Birth Weight (Grams) | 1891.8 ± 935.5 | 2093.7 ± 1136.9 |
| Female (%) | 38.5% | 37.9% |
| Premature (GA < 37 Weeks) (%) | 76.9% | 62.1% |
| Group 1 Neonates | ||
| Patient | Main Diagnosis | Patient Outcome |
| P1 | Premature | Discharge |
| P2 | Premature, patent ductus arteriosus (PDA) with surgery, posthemorrhagic ventriculomegaly | Discharge |
| P3 | Premature, intestinal volvulus with surgery | Discharge |
| P4 | Polymalformative syndrome | Discharge |
| P5 | Premature | Discharge |
| P6 | Premature, pulmonary hypertension (PH), stroke | Discharge |
| P7 | Premature, bronchopulmonary dysplasia (BPD) | Discharge |
| P9 | Congenital diaphragmatic hernia (CDH) | Discharge |
| P10 | Epidermolysis | Discharge |
| P15 | Congenital myopathy | Discharge |
| P17 | Congenital cardiopathy, extracorporeal membrane oxygenation (ECMO) | Exitus |
| P18 | Premature, intestinal perforation, chylothorax | Exitus |
| P19 | Congenital atrioventricular block | Discharge |
| Group 2 Neonates | ||
| Patient | Main Diagnosis | Patient Outcome |
| P20 | Oesophageal atresia | Discharge |
| P21 | Premature, PDA with surgery, PH | Discharge |
| P22 | Premature, PDA with surgery, BPD, extensive leukomalacia | Discharge |
| P23 | Congenital hyperinsulinism | Discharge |
| P24 | Premature, Pulmonary Hypoplasia, BPD | Discharge |
| P25 | Noonan Syndrome, hypertrophic cardiomyopathy | Exitus |
| P26 | Congenital heart disease: Transposition of the Great Arteries (TGA) | Discharge |
| P27 | Congenital chylothorax | Exitus |
| P28 | Congenital heart disease: total pulmonary venous drainage, kidney failure | Exitus |
| P29 | Hyaline membrane disease, pneumothorax | Discharge |
| P30 | CDH, PH | Discharge |
| P31 | Premature, BPD | Discharge |
| P32 | Premature, PH, posthemorrhagic ventriculomegalia | Discharge |
| P33 | Premature | Discharge |
| P34 | CDH, PH | Exitus |
| P35 | Premature, BPD | Discharge |
| P36 | Oesophageal atresia | Discharge |
| P37 | Premature | Discharge |
| P38 | Premature, congenital tuberculosis | Discharge |
| P39 | Congenital hyperinsulinism | Discharge |
| P40 | Premature, BPD | Discharge |
| P41 | Transfer at 3 months of life, premature, BPD, necrotizing enterocolitis (NEC) | Discharge |
| P42 | Premature | Discharge |
| P43 | Premature | Discharge |
| P44 | Congenital heart disease: Tetralogy of Fallot | Discharge |
| P45 | Congenital heart disease: total pulmonary venous drainage, kidney failure | Discharge |
| P46 | Hirschsprung disease, intestinal perforation, ileostomy | Discharge |
| P47 | Premature | Discharge |
| P48 | Oesophageal atresia | Discharge |
| Patient | Gender | GA (Weeks) | Age (Days) | Prior Colonization | Previous Antibiotics | Extraintestinal Serratia spp. |
|---|---|---|---|---|---|---|
| P1 | Female | 32.1 | 13 | No | No | Conjuctiva (conjunctivitis) |
| P2 | Male | 26.5 | 27 | No | Yes | Bronchial aspirate (pneumonia) |
| P3 | Male | 32.6 | 14 | Yes | Yes | Blood, cerebrospinal fluid, skin wound, and skin around ileostomy (sepsis and meningitis) |
| P4 | Male | 37.6 | 11 | ND | Yes | None |
| P5 | Male | 26 | 27 | Yes | Yes | Blood (bacteriemia) |
| P6 | Male | 27.5 | 24 | No | Yes | Conjuctiva (two isolates) (conjunctivitis) |
| P7 | Female | 29.4 | 18 | ND | Yes | None |
| P9 | Female | 37.2 | 14 | ND | Yes | None |
| P10 | Female | 35 | 15 | ND | Yes | None |
| P15 | Male | 36.5 | 29 | ND | No | None |
| P17 | Male | 38 | 11 | Yes | Yes | Bronchial aspirate (colonization) |
| P18 | Female | 24.4 | 17 | Yes | Yes | Blood (two isolates), bronchial aspirate, pericatheter skin, and skin around ileostomy (sepsis) |
| P19 | Male | 35.2 | 7 | No | Yes | Blood (sepsis) |
| Strain | Gene (Antibiotic Resistance) | |||||
|---|---|---|---|---|---|---|
| qnrB56 (Quinolones) | aac(6′)-Ic (Aminoglycosides) | blaSRT-2 (β-Lactams) | blaSST-1 (β-Lactams) | oqxB (Efflux Pump) | tetA41 (Tetracycline) | |
| S1 | ✕ | ✓ | ✕ | ✓ | ✓ | ✓ |
| S3B | ✕ | ✓ | ✕ | ✓ | ✓ | ✓ |
| S9 | ✕ | ✓ | ✕ | ✓ | ✓ | ✓ |
| S17 | ✓ | ✓ | ✓ | ✕ | ✓ | ✕ |
| S18 | ✓ | ✓ | ✓ | ✕ | ✓ | ✓ |
| S19 | ✓ | ✓ | ✓ | ✕ | ✓ | ✓ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dahdouh, E.; Lázaro-Perona, F.; Ruiz-Carrascoso, G.; Sánchez García, L.; Saenz de Pipaón, M.; Mingorance, J. Intestinal Dominance by Serratia marcescens and Serratia ureilytica among Neonates in the Setting of an Outbreak. Microorganisms 2021, 9, 2271. https://doi.org/10.3390/microorganisms9112271
Dahdouh E, Lázaro-Perona F, Ruiz-Carrascoso G, Sánchez García L, Saenz de Pipaón M, Mingorance J. Intestinal Dominance by Serratia marcescens and Serratia ureilytica among Neonates in the Setting of an Outbreak. Microorganisms. 2021; 9(11):2271. https://doi.org/10.3390/microorganisms9112271
Chicago/Turabian StyleDahdouh, Elias, Fernando Lázaro-Perona, Guillermo Ruiz-Carrascoso, Laura Sánchez García, Miguel Saenz de Pipaón, and Jesús Mingorance. 2021. "Intestinal Dominance by Serratia marcescens and Serratia ureilytica among Neonates in the Setting of an Outbreak" Microorganisms 9, no. 11: 2271. https://doi.org/10.3390/microorganisms9112271
APA StyleDahdouh, E., Lázaro-Perona, F., Ruiz-Carrascoso, G., Sánchez García, L., Saenz de Pipaón, M., & Mingorance, J. (2021). Intestinal Dominance by Serratia marcescens and Serratia ureilytica among Neonates in the Setting of an Outbreak. Microorganisms, 9(11), 2271. https://doi.org/10.3390/microorganisms9112271







