The SPFH Protein Superfamily in Fungi: Impact on Mitochondrial Function and Implications in Virulence
Abstract
:1. Introduction
2. SPFH Protein Function in Non-Pathogenic Fungi
3. SPFH Protein Function in Pathogenic Fungi
4. Perspectives and Future Directions
Author Contributions
Funding
Conflicts of Interest
References
- Lapatsina, L.; Brand, J.; Poole, K.; Daumke, O.; Lewin, G.R. Stomatin-domain proteins. Eur. J. Cell Biol. 2012, 91, 240–245. [Google Scholar] [CrossRef]
- Browman, D.T.; Hoegg, M.B.; Robbins, S.M. The SPFH domain-containing proteins: More than lipid raft markers. Trends Cell Biol. 2007, 17, 394–402. [Google Scholar] [CrossRef]
- Rivera-Milla, E.; Stuermer, C.A.; Malaga-Trillo, E. Ancient origin of reggie (flotillin), reggie-like, and other lipid-raft proteins: Convergent evolution of the SPFH domain. Cell. Mol. Life Sci. 2006, 63, 343–357. [Google Scholar] [CrossRef] [Green Version]
- Hinderhofer, M.; Walker, C.A.; Friemel, A.; Stuermer, C.A.; Moller, H.M.; Reuter, A. Evolution of prokaryotic SPFH proteins. BMC Evol. Biol. 2009, 9, 10. [Google Scholar] [CrossRef] [Green Version]
- Conrad, K.A.; Rodriguez, R.; Salcedo, E.C.; Rauceo, J.M. The Candida albicans stress response gene Stomatin-Like Protein 3 is implicated in ROS-induced apoptotic-like death of yeast phase cells. PLoS ONE 2018, 13, e0192250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cabezon, V.; Llama-Palacios, A.; Nombela, C.; Monteoliva, L.; Gil, C. Analysis of Candida albicans plasma membrane proteome. Proteomics 2009, 9, 4770–4786. [Google Scholar] [CrossRef] [PubMed]
- Snyers, L.; Umlauf, E.; Prohaska, R. Oligomeric nature of the integral membrane protein stomatin. J. Biol. Chem. 1998, 273, 17221–17226. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hiebl-Dirschmied, C.M.; Adolf, G.R.; Prohaska, R. Isolation and partial characterization of the human erythrocyte band 7 integral membrane protein. Biochim. Biophys. Acta 1991, 1065, 195–202. [Google Scholar] [CrossRef]
- Tatsuta, T.; Model, K.; Langer, T. Formation of membrane-bound ring complexes by prohibitins in mitochondria. Mol. Biol. Cell 2005, 16, 248–259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Da Cruz, S.; Parone, P.A.; Gonzalo, P.; Bienvenut, W.V.; Tondera, D.; Jourdain, A.; Quadroni, M.; Martinou, J.C. SLP-2 interacts with prohibitins in the mitochondrial inner membrane and contributes to their stability. Biochim. Biophys. Acta 2008, 1783, 904–911. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Christie, D.A.; Kirchhof, M.G.; Vardhana, S.; Dustin, M.L.; Madrenas, J. Mitochondrial and plasma membrane pools of stomatin-like protein 2 coalesce at the immunological synapse during T cell activation. PLoS ONE 2012, 7, e37144. [Google Scholar] [CrossRef] [Green Version]
- Dempwolff, F.; Schmidt, F.K.; Hervas, A.B.; Stroh, A.; Rosch, T.C.; Riese, C.N.; Dersch, S.; Heimerl, T.; Lucena, D.; Hulsbusch, N.; et al. Super Resolution Fluorescence Microscopy and Tracking of Bacterial Flotillin (Reggie) Paralogs Provide Evidence for Defined-Sized Protein Microdomains within the Bacterial Membrane but Absence of Clusters Containing Detergent-Resistant Proteins. PLoS Genet. 2016, 12, e1006116. [Google Scholar] [CrossRef] [PubMed]
- Reuter, A.T.; Stuermer, C.A.; Plattner, H. Identification, localization, and functional implications of the microdomain-forming stomatin family in the ciliated protozoan Paramecium tetraurelia. Eukaryot. Cell 2013, 12, 529–544. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mairhofer, M.; Steiner, M.; Salzer, U.; Prohaska, R. Stomatin-like protein-1 interacts with stomatin and is targeted to late endosomes. J. Biol. Chem. 2009, 284, 29218–29229. [Google Scholar] [CrossRef] [Green Version]
- Wai, T.; Saita, S.; Nolte, H.; Muller, S.; Konig, T.; Richter-Dennerlein, R.; Sprenger, H.G.; Madrenas, J.; Muhlmeister, M.; Brandt, U.; et al. The membrane scaffold SLP2 anchors a proteolytic hub in mitochondria containing PARL and the i-AAA protease YME1L. EMBO Rep. 2016, 17, 1844–1856. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rungaldier, S.; Umlauf, E.; Mairhofer, M.; Salzer, U.; Thiele, C.; Prohaska, R. Structure-function analysis of human stomatin: A mutation study. PLoS ONE 2017, 12, e0178646. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yokoyama, H.; Matsui, I. Crystal structure of the stomatin operon partner protein from Pyrococcus horikoshii indicates the formation of a multimeric assembly. FEBS Open Bio 2014, 4, 804–812. [Google Scholar] [CrossRef] [Green Version]
- Wei, Y.; Chiang, W.C.; Sumpter, R., Jr.; Mishra, P.; Levine, B. Prohibitin 2 Is an Inner Mitochondrial Membrane Mitophagy Receptor. Cell 2017, 168, 224–238. [Google Scholar] [CrossRef] [Green Version]
- Yan, C.; Gong, L.; Chen, L.; Xu, M.; Abou-Hamdan, H.; Tang, M.; Desaubry, L.; Song, Z. PHB2 (prohibitin 2) promotes PINK1-PRKN/Parkin-dependent mitophagy by the PARL-PGAM5-PINK1 axis. Autophagy 2020, 16, 419–434. [Google Scholar] [CrossRef]
- Mitsopoulos, P.; Lapohos, O.; Weraarpachai, W.; Antonicka, H.; Chang, Y.H.; Madrenas, J. Stomatin-like protein 2 deficiency results in impaired mitochondrial translation. PLoS ONE 2017, 12, e0179967. [Google Scholar] [CrossRef] [Green Version]
- Christie, D.A.; Lemke, C.D.; Elias, I.M.; Chau, L.A.; Kirchhof, M.G.; Li, B.; Ball, E.H.; Dunn, S.D.; Hatch, G.M.; Madrenas, J. Stomatin-like protein 2 binds cardiolipin and regulates mitochondrial biogenesis and function. Mol. Cell. Biol. 2011, 31, 3845–3856. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mitsopoulos, P.; Chang, Y.H.; Wai, T.; Konig, T.; Dunn, S.D.; Langer, T.; Madrenas, J. Stomatin-like protein 2 is required for in vivo mitochondrial respiratory chain supercomplex formation and optimal cell function. Mol. Cell. Biol. 2015, 35, 1838–1847. [Google Scholar] [CrossRef] [Green Version]
- Klipp, R.C.; Cullinan, M.M.; Bankston, J.R. Insights into the molecular mechanisms underlying the inhibition of acid-sensing ion channel 3 gating by stomatin. J. Gen. Physiol. 2020, 152, e201912471. [Google Scholar] [CrossRef] [PubMed]
- Wetzel, C.; Hu, J.; Riethmacher, D.; Benckendorff, A.; Harder, L.; Eilers, A.; Moshourab, R.; Kozlenkov, A.; Labuz, D.; Caspani, O.; et al. A stomatin-domain protein essential for touch sensation in the mouse. Nature 2007, 445, 206–209. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.; Gu, G.; Ferguson, E.L.; Chalfie, M. A stomatin-like protein necessary for mechanosensation in C. elegans. Nature 1995, 378, 292–295. [Google Scholar] [CrossRef]
- Chowdhury, I.; Thompson, W.E.; Thomas, K. Prohibitins role in cellular survival through Ras-Raf-MEK-ERK pathway. J. Cell. Physiol. 2014, 229, 998–1004. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bartolome, A.; Boskovic, S.; Paunovic, I.; Bozic, V.; Cvejic, D. Stomatin-like protein 2 overexpression in papillary thyroid carcinoma is significantly associated with high-risk clinicopathological parameters and BRAFV600E mutation. APMIS 2016, 124, 271–277. [Google Scholar] [CrossRef]
- Signorile, A.; Sgaramella, G.; Bellomo, F.; De Rasmo, D. Prohibitins: A Critical Role in Mitochondrial Functions and Implication in Diseases. Cells 2019, 8, 71. [Google Scholar] [CrossRef] [Green Version]
- Zhu, W.; Li, W.; Geng, Q.; Wang, X.; Sun, W.; Jiang, H.; Pu, X. Silence of Stomatin-Like Protein 2 Represses Migration and Invasion Ability of Human Liver Cancer Cells via Inhibiting the Nuclear Factor Kappa B (NF-kappaB) Pathway. Med. Sci. Monit. 2018, 24, 7625–7632. [Google Scholar] [CrossRef]
- Matz, J.M.; Goosmann, C.; Matuschewski, K.; Kooij, T.W.A. An Unusual Prohibitin Regulates Malaria Parasite Mitochondrial Membrane Potential. Cell Rep. 2018, 23, 756–767. [Google Scholar] [CrossRef] [Green Version]
- Cruz-Bustos, T.; Ibarrola-Vannucci, A.K.; Diaz-Lozano, I.; Ramirez, J.L.; Osuna, A. Characterization and functionality of two members of the SPFH protein superfamily, prohibitin 1 and 2 in Leishmania major. Parasit. Vectors 2018, 11, 622. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tyc, J.; Faktorova, D.; Kriegova, E.; Jirku, M.; Vavrova, Z.; Maslov, D.A.; Lukes, J. Probing for primary functions of prohibitin in Trypanosoma brucei. Int. J. Parasitol. 2010, 40, 73–83. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Yao, F.; Jiang, G.; Xu, M.; Chen, S.; Zhou, L.; Sakamoto, N.; Kuno, T.; Fang, Y. Dysfunction of Prohibitin 2 Results in Reduced Susceptibility to Multiple Antifungal Drugs via Activation of the Oxidative Stress-Responsive Transcription Factor Pap1 in Fission Yeast. Antimicrob. Agents Chemother. 2018, 62, e00860-18. [Google Scholar] [CrossRef] [Green Version]
- Klecker, T.; Wemmer, M.; Haag, M.; Weig, A.; Bockler, S.; Langer, T.; Nunnari, J.; Westermann, B. Interaction of MDM33 with mitochondrial inner membrane homeostasis pathways in yeast. Sci. Rep. 2015, 5, 18344. [Google Scholar] [CrossRef] [PubMed]
- Sikora, J.; Towpik, J.; Graczyk, D.; Kistowski, M.; Rubel, T.; Poznanski, J.; Langridge, J.; Hughes, C.; Dadlez, M.; Boguta, M. Yeast prion [PSI+] lowers the levels of mitochondrial prohibitins. Biochim. Biophys. Acta 2009, 1793, 1703–1709. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Osman, C.; Haag, M.; Potting, C.; Rodenfels, J.; Dip, P.V.; Wieland, F.T.; Brugger, B.; Westermann, B.; Langer, T. The genetic interactome of prohibitins: Coordinated control of cardiolipin and phosphatidylethanolamine by conserved regulators in mitochondria. J. Cell Biol. 2009, 184, 583–596. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Osman, C.; Wilmes, C.; Tatsuta, T.; Langer, T. Prohibitins interact genetically with Atp23, a novel processing peptidase and chaperone for the F1Fo-ATP synthase. Mol. Biol. Cell 2007, 18, 627–635. [Google Scholar] [CrossRef] [Green Version]
- Marques, I.; Dencher, N.A.; Videira, A.; Krause, F. Supramolecular organization of the respiratory chain in Neurospora crassa mitochondria. Eukaryot. Cell 2007, 6, 2391–2405. [Google Scholar] [CrossRef] [Green Version]
- Kirchman, P.A.; Miceli, M.V.; West, R.L.; Jiang, J.C.; Kim, S.; Jazwinski, S.M. Prohibitins and Ras2 protein cooperate in the maintenance of mitochondrial function during yeast aging. Acta Biochim. Pol. 2003, 50, 1039–1056. [Google Scholar] [CrossRef] [Green Version]
- Piper, P.W.; Jones, G.W.; Bringloe, D.; Harris, N.; MacLean, M.; Mollapour, M. The shortened replicative life span of prohibitin mutants of yeast appears to be due to defective mitochondrial segregation in old mother cells. Aging Cell 2002, 1, 149–157. [Google Scholar] [CrossRef] [Green Version]
- Glynn, S.E. Multifunctional Mitochondrial AAA Proteases. Front. Mol. Biosci. 2017, 4, 34. [Google Scholar] [CrossRef] [Green Version]
- Calderone, R.; Li, D.; Traven, A. System-level impact of mitochondria on fungal virulence: To metabolism and beyond. FEMS Yeast Res. 2015, 15, fov027. [Google Scholar] [CrossRef] [Green Version]
- Mamouei, Z.; Singh, S.; Lemire, B.; Gu, Y.; Alqarihi, A.; Nabeela, S.; Li, D.; Ibrahim, A.; Uppuluri, P. An evolutionarily diverged mitochondrial protein controls biofilm growth and virulence in Candida albicans. PLoS Biol. 2021, 19, e3000957. [Google Scholar] [CrossRef]
- Koch, B.; Traven, A. Mitochondrial Control of Fungal Cell Walls: Models and Relevance in Fungal Pathogens. Curr. Top. Microbiol. Immunol. 2020, 425, 277–296. [Google Scholar] [CrossRef] [PubMed]
- Richard, A.; Calderone, C.J.C. (Eds.) Candida and Candidiasis, 2nd ed.; ASM Press: Washington, DC, USA, 2012; p. 524. [Google Scholar]
- Sobel, J.D. Recurrent vulvovaginal candidiasis. Am. J. Obstet. Gynecol. 2016, 214, 15–21. [Google Scholar] [CrossRef]
- Brown, G.D.; Denning, D.W.; Gow, N.A.; Levitz, S.M.; Netea, M.G.; White, T.C. Hidden killers: Human fungal infections. Sci. Transl. Med. 2012, 4, 165rv113. [Google Scholar] [CrossRef] [Green Version]
- Pfaller, M.A.; Diekema, D.J. Epidemiology of invasive candidiasis: A persistent public health problem. Clin. Microbiol. Rev. 2007, 20, 133–163. [Google Scholar] [CrossRef] [Green Version]
- Duvenage, L.; Walker, L.A.; Bojarczuk, A.; Johnston, S.A.; MacCallum, D.M.; Munro, C.A.; Gourlay, C.W. Inhibition of Classical and Alternative Modes of Respiration in Candida albicans Leads to Cell Wall Remodeling and Increased Macrophage Recognition. mBio 2019, 10, e02535-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koch, B.; Tucey, T.M.; Lo, T.L.; Novakovic, S.; Boag, P.; Traven, A. The Mitochondrial GTPase Gem1 Contributes to the Cell Wall Stress Response and Invasive Growth of Candida albicans. Front. Microbiol. 2017, 8, 2555. [Google Scholar] [CrossRef]
- Li, D.; Chen, H.; Florentino, A.; Alex, D.; Sikorski, P.; Fonzi, W.A.; Calderone, R. Enzymatic dysfunction of mitochondrial complex I of the Candida albicans goa1 mutant is associated with increased reactive oxidants and cell death. Eukaryot. Cell 2011, 10, 672–682. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bambach, A.; Fernandes, M.P.; Ghosh, A.; Kruppa, M.; Alex, D.; Li, D.; Fonzi, W.A.; Chauhan, N.; Sun, N.; Agrellos, O.A.; et al. Goa1p of Candida albicans localizes to the mitochondria during stress and is required for mitochondrial function and virulence. Eukaryot. Cell 2009, 8, 1706–1720. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, N.; Parrish, R.S.; Calderone, R.A.; Fonzi, W.A. Unique, Diverged, and Conserved Mitochondrial Functions Influencing Candida albicans Respiration. mBio 2019, 10, e00300-19. [Google Scholar] [CrossRef] [Green Version]
- Heredia, M.Y.; Ikeh, M.A.C.; Gunasekaran, D.; Conrad, K.A.; Filimonava, S.; Marotta, D.H.; Nobile, C.J.; Rauceo, J.M. An expanded cell wall damage signaling network is comprised of the transcription factors Rlm1 and Sko1 in Candida albicans. PLoS Genet. 2020, 16, e1008908. [Google Scholar] [CrossRef]
- Marotta, D.H.; Nantel, A.; Sukala, L.; Teubl, J.R.; Rauceo, J.M. Genome-wide transcriptional profiling and enrichment mapping reveal divergent and conserved roles of Sko1 in the Candida albicans osmotic stress response. Genomics 2013, 102, 363–371. [Google Scholar] [CrossRef]
- Rauceo, J.M.; Blankenship, J.R.; Fanning, S.; Hamaker, J.J.; Deneault, J.S.; Smith, F.J.; Nantel, A.; Mitchell, A.P. Regulation of the Candida albicans cell wall damage response by transcription factor Sko1 and PAS kinase Psk1. Mol. Biol. Cell 2008, 19, 2741–2751. [Google Scholar] [CrossRef] [Green Version]
- Duvenage, L.; Munro, C.A.; Gourlay, C.W. The potential of respiration inhibition as a new approach to combat human fungal pathogens. Curr. Genet. 2019, 65, 1347–1353. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Joseph-Horne, T.; Hollomon, D.W.; Wood, P.M. Fungal respiration: A fusion of standard and alternative components. Biochim. Biophys. Acta 2001, 1504, 179–195. [Google Scholar] [CrossRef] [Green Version]
- Skrzypek, M.S.; Binkley, J.; Binkley, G.; Miyasato, S.R.; Simison, M.; Sherlock, G. The Candida Genome Database (CGD): Incorporation of Assembly 22, systematic identifiers and visualization of high throughput sequencing data. Nucleic Acids Res. 2017, 45, D592–D596. [Google Scholar] [CrossRef] [Green Version]
- Wetzel, C.; Pifferi, S.; Picci, C.; Gok, C.; Hoffmann, D.; Bali, K.K.; Lampe, A.; Lapatsina, L.; Fleischer, R.; Smith, E.S.; et al. Small-molecule inhibition of STOML3 oligomerization reverses pathological mechanical hypersensitivity. Nat. Neurosci. 2017, 20, 209–218. [Google Scholar] [CrossRef] [PubMed]
- Nantel, A.; Dignard, D.; Bachewich, C.; Harcus, D.; Marcil, A.; Bouin, A.P.; Sensen, C.W.; Hogues, H.; van het Hoog, M.; Gordon, P.; et al. Transcription profiling of Candida albicans cells undergoing the yeast-to-hyphal transition. Mol. Biol. Cell 2002, 13, 3452–3465. [Google Scholar] [CrossRef] [Green Version]
- Chen, T.W.; Liu, H.W.; Liou, Y.J.; Lee, J.H.; Lin, C.H. Over-expression of stomatin causes syncytium formation in nonfusogenic JEG-3 choriocarcinoma placental cells. Cell Biol. Int. 2016, 40, 926–933. [Google Scholar] [CrossRef]
- Takeshita, N.; Diallinas, G.; Fischer, R. The role of flotillin FloA and stomatin StoA in the maintenance of apical sterol-rich membrane domains and polarity in the filamentous fungus Aspergillus nidulans. Mol. Microbiol. 2012, 83, 1136–1152. [Google Scholar] [CrossRef]
- Narasimhan, S.; Armstrong, M.; McClung, J.K.; Richards, F.F.; Spicer, E.K. Prohibitin, a putative negative control element present in Pneumocystis carinii. Infect. Immun. 1997, 65, 5125–5130. [Google Scholar] [CrossRef] [Green Version]
- Lock, A.; Rutherford, K.; Harris, M.A.; Hayles, J.; Oliver, S.G.; Bahler, J.; Wood, V. PomBase 2018: User-driven reimplementation of the fission yeast database provides rapid and intuitive access to diverse, interconnected information. Nucleic Acids Res. 2019, 47, D821–D827. [Google Scholar] [CrossRef] [Green Version]
- Cherry, J.M.; Hong, E.L.; Amundsen, C.; Balakrishnan, R.; Binkley, G.; Chan, E.T.; Christie, K.R.; Costanzo, M.C.; Dwight, S.S.; Engel, S.R.; et al. Saccharomyces Genome Database: The genomics resource of budding yeast. Nucleic Acids Res. 2012, 40, D700–D705. [Google Scholar] [CrossRef] [Green Version]
- Jones, P.; Binns, D.; Chang, H.Y.; Fraser, M.; Li, W.; McAnulla, C.; McWilliam, H.; Maslen, J.; Mitchell, A.; Nuka, G.; et al. InterProScan 5: Genome-scale protein function classification. Bioinformatics 2014, 30, 1236–1240. [Google Scholar] [CrossRef] [Green Version]
- Liu, W.; Xie, Y.; Ma, J.; Luo, X.; Nie, P.; Zuo, Z.; Lahrmann, U.; Zhao, Q.; Zheng, Y.; Zhao, Y.; et al. IBS: An illustrator for the presentation and visualization of biological sequences. Bioinformatics 2015, 31, 3359–3361. [Google Scholar] [CrossRef] [Green Version]
- Li, D.; Calderone, R. Exploiting mitochondria as targets for the development of new antifungals. Virulence 2017, 8, 159–168. [Google Scholar] [CrossRef] [Green Version]
- Berman, J.; Krysan, D.J. Drug resistance and tolerance in fungi. Nat. Rev. Microbiol. 2020, 18, 319–331. [Google Scholar] [CrossRef]
- Benhamou, R.I.; Bibi, M.; Steinbuch, K.B.; Engel, H.; Levin, M.; Roichman, Y.; Berman, J.; Fridman, M. Real-Time Imaging of the Azole Class of Antifungal Drugs in Live Candida Cells. ACS Chem. Biol. 2017, 12, 1769–1777. [Google Scholar] [CrossRef]
- Wang, D.; Tabti, R.; Elderwish, S.; Djehal, A.; Chouha, N.; Pinot, F.; Yu, P.; Nebigil, C.G.; Desaubry, L. SFPH proteins as therapeutic targets for a myriad of diseases. Bioorg. Med. Chem. Lett. 2020, 30, 127600. [Google Scholar] [CrossRef] [PubMed]
- Westermann, B. The mitochondria-plasma membrane contact site. Curr. Opin. Cell Biol. 2015, 35, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, N.; Quail, M.M.F.; Hernday, A.D. An Efficient, Rapid, and Recyclable System for CRISPR-Mediated Genome Editing in Candida albicans. mSphere 2017, 2, e00149-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rungaldier, S.; Oberwagner, W.; Salzer, U.; Csaszar, E.; Prohaska, R. Stomatin interacts with GLUT1/SLC2A1, band 3/SLC4A1, and aquaporin-1 in human erythrocyte membrane domains. Biochim. Biophys. Acta 2013, 1828, 956–966. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pankow, S.; Bamberger, C.; Calzolari, D.; Bamberger, A.; Yates, J.R., 3rd. Deep interactome profiling of membrane proteins by co-interacting protein identification technology. Nat. Protoc. 2016, 11, 2515–2528. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maciel, E.I.; Jiang, C.; Barghouth, P.G.; Nobile, C.J.; Oviedo, N.J. The planarian Schmidtea mediterranea is a new model to study host-pathogen interactions during fungal infections. Dev. Comp. Immunol. 2019, 93, 18–27. [Google Scholar] [CrossRef] [Green Version]


| Protein | Localization | Function | References |
|---|---|---|---|
| S. cerevisiae | |||
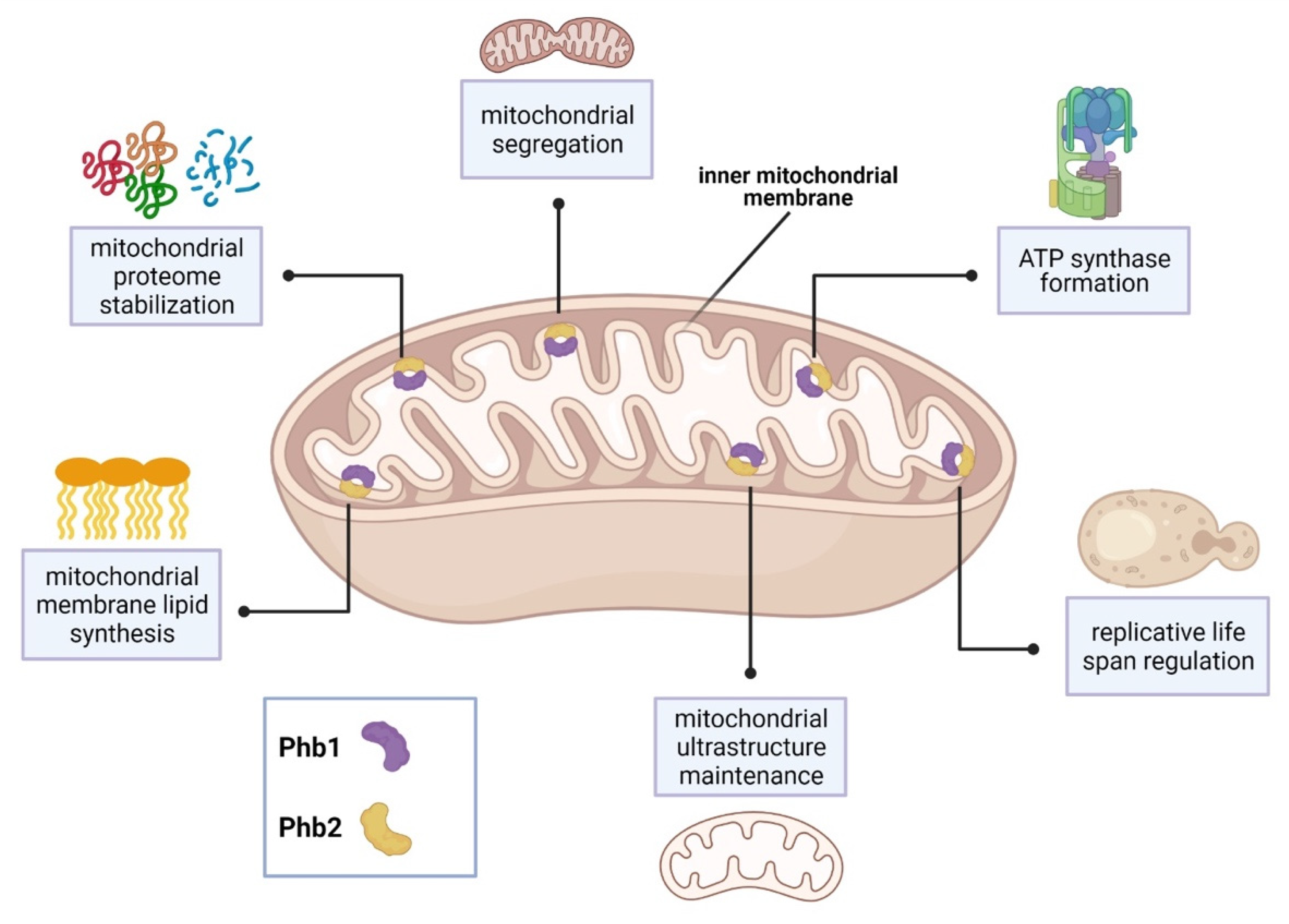
| Phb1 and Phb2 | inner mitochondrial membrane, Phb1-Phb2 complex | regulation of mitochondrial ultrastructure and segregation, mitochondrial protein stabilization, regulation of replicative life span, regulation of mitochondrial membrane lipid synthesis, involvement in ATP synthase formation | [34,35,36,37,39,40] |
| S. pombe | |||
| Phb1 | mitochondria | multi-drug resistance | [33] |
| Phb2 | mitochondria | multi-drug resistance, oxidative stress signaling | [33] |
| C. albicans | |||
| Slp3 | plasma membrane, vacuolar lumen | yeast-specific general stress response signaling, involved in maintenance of mitochondrial membrane integrity | [5,6] |
| N. crassa | |||
| Slp2 | mitochondria | interactions with i-MMM protein IAP-1 | [38] |
| Phb1 and Phb2 | inner mitochondrial membrane, Phb1-Phb2 complex | interactions with m-AAA protease MAP-1 | [38] |
| A. nidulans | |||
| FloA | plasma membrane | formation of sterol-rich domains in plasma membrane | [63] |
| StoA | plasma membrane, endosome/vacuole | polarized hyphal growth | [63] |
| P. carinii | |||
| Prohibitin | inner mitochondrial membrane (predicted) | regulation of cell proliferation and development | [64] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Heredia, M.Y.; Rauceo, J.M. The SPFH Protein Superfamily in Fungi: Impact on Mitochondrial Function and Implications in Virulence. Microorganisms 2021, 9, 2287. https://doi.org/10.3390/microorganisms9112287
Heredia MY, Rauceo JM. The SPFH Protein Superfamily in Fungi: Impact on Mitochondrial Function and Implications in Virulence. Microorganisms. 2021; 9(11):2287. https://doi.org/10.3390/microorganisms9112287
Chicago/Turabian StyleHeredia, Marienela Y., and Jason M. Rauceo. 2021. "The SPFH Protein Superfamily in Fungi: Impact on Mitochondrial Function and Implications in Virulence" Microorganisms 9, no. 11: 2287. https://doi.org/10.3390/microorganisms9112287






