Periodontal Pathogens Inhabit Root Caries Lesions Extending beyond the Gingival Margin: A Next-Generation Sequencing Analysis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Population
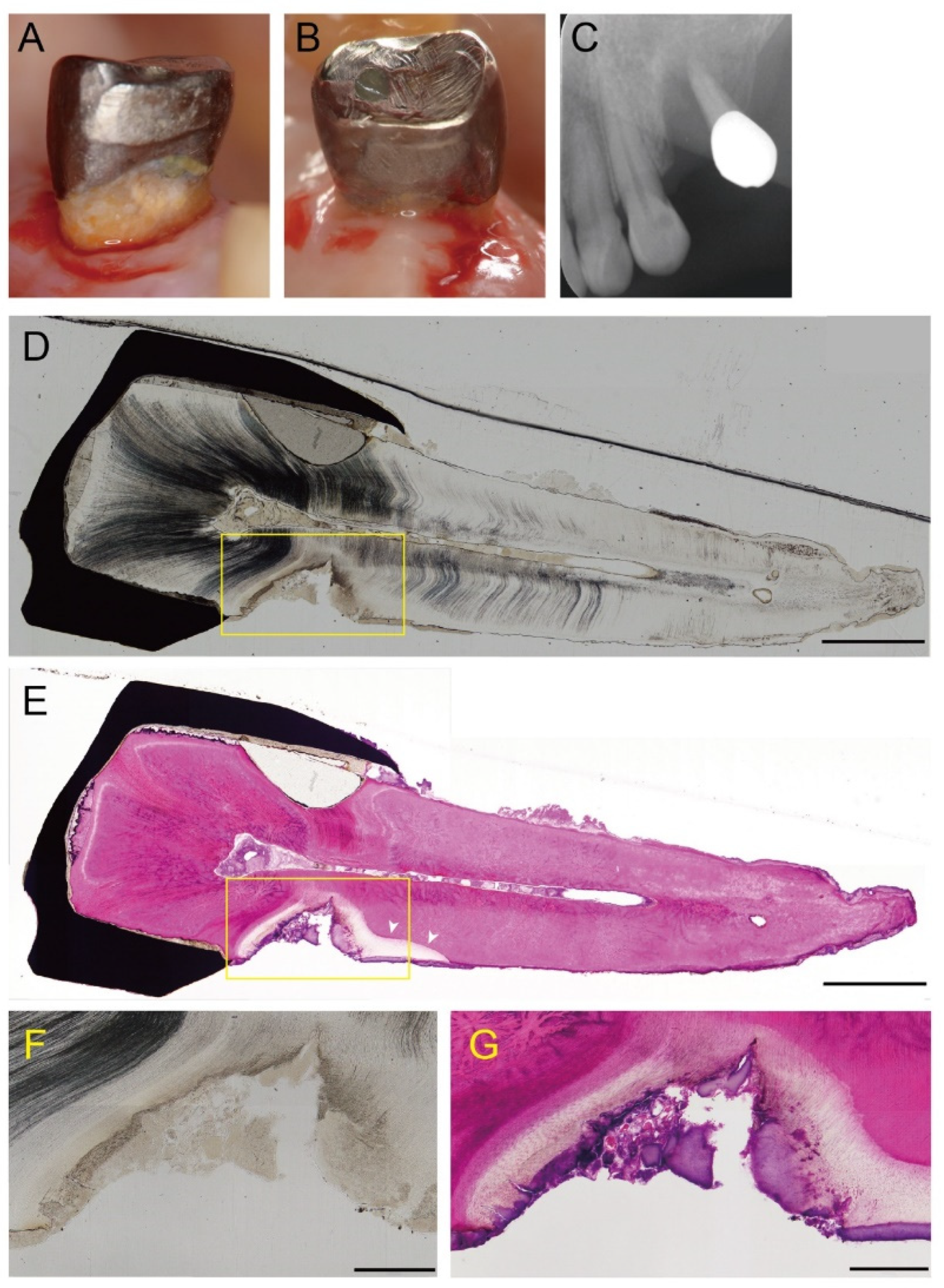
2.2. Histological Observation
2.3. Sample Collection and DNA Extraction
2.4. Microbiome Analysis
2.5. Statistical Analysis
3. Results
3.1. Demographic and Clinical Characteristics
3.2. Histopathology of Root Caries
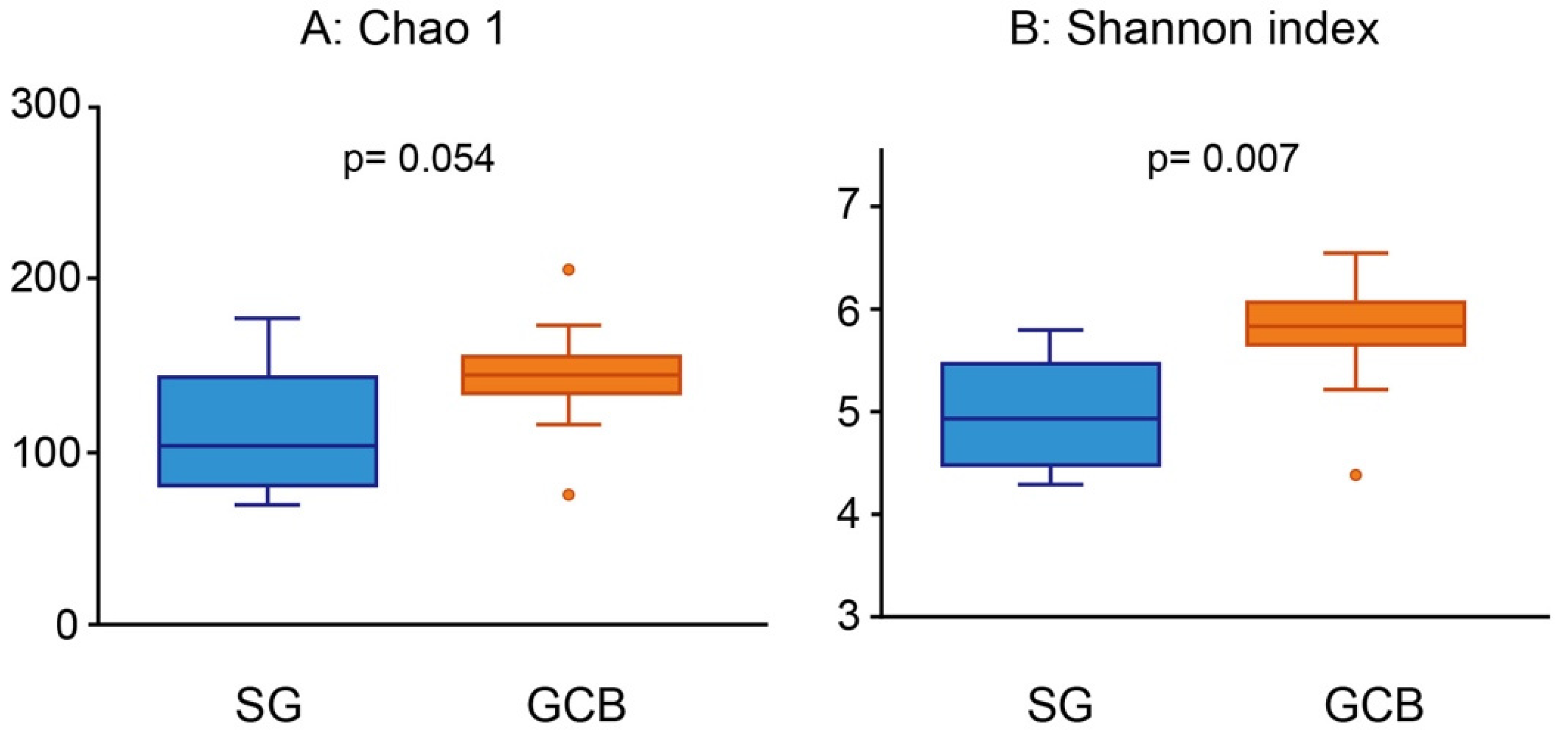
3.3. Alpha Diversity Analyses of the Bacterial Community
3.4. Differentiating Composition of SG and GCB
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Griffin, S.O.; Griffin, P.M.; Swann, J.L.; Zlobin, N. Estimating rates of new root caries in older adults. J. Dent. Res. 2004, 83, 634–638. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.B.; Hu, T.; Zhou, X.D.; Shao, R.; Cheng, R.; Wang, G.S.; Yang, Y.M.; Li, X.; Yuan, B.; Xu, T.; et al. How root caries differs between middle-aged people and the elderly: Findings from the 4th national oral health survey of China. Chin. J. Dent. Res. 2018, 21, 221–229. [Google Scholar] [PubMed]
- Hariyani, N.; Setyowati, D.; Spencer, A.J.; Luzzi, L.; Do, L.G. Root caries incidence and increment in the population—A systematic review, meta-analysis and meta-regression of longitudinal studies. J. Dent. 2018, 77, 1–7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, J.; Leung, K.C.M.; Sardana, D.; Wong, M.C.M.; Lo, E.C.M. Risk predictors of dental root caries: A systematic review. J. Dent. 2019, 89, 103166. [Google Scholar] [CrossRef]
- Paris, S.; Banerjee, A.; Bottenberg, P.; Breschi, L.; Campus, G.; Doméjean, S.; Ekstrand, K.; Giacaman, R.A.; Haak, R.; Hanning, M.; et al. How to intervene in the caries process in older adults: A joint ORCA and EFCD expert Delphi consensus statement. Caries Res. 2020, 54, 459–465. [Google Scholar] [CrossRef]
- Bignozzi, I.; Crea, A.; Capri, D.; Littarru, C.; Lajolo, C.; Tatakis, D.N. Root caries: A periodontal perspective. J. Periodont. Res. 2014, 49, 143–163. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, N.; Nyvad, B. Ecological hypothesis of dentin and root caries. Caries Res. 2016, 50, 422–431. [Google Scholar] [CrossRef] [PubMed]
- Keltjens, H.M.A.; Schaeken, M.J.M.; van der Hoeven, J.S.; Hendricks, J.C.M. Microflora of plaque from sound and carious root surfaces. Caries Res. 1987, 21, 193–199. [Google Scholar] [CrossRef] [PubMed]
- van Houte, J.; Lopman, J.; Kent, R. The final pH of bacteria comprising the predominant flora on sound and carious human root and enamel surfaces. J. Dent. Res. 1996, 75, 1008–1014. [Google Scholar] [CrossRef] [PubMed]
- Schüpbach, P.; Osterwalder, V.; Guggenheim, B. Human root caries: Microbiota of a limited number of root caries lesions. Caries Res. 1996, 30, 52–64. [Google Scholar] [CrossRef]
- Brailsford, S.R.; Tregaskis, R.B.; Leftwich, H.S.; Beighton, D. The predominant Actinomyces spp. Isolated from infected dentin of active root caries lesions. J. Dent. Res. 1999, 78, 1525–1534. [Google Scholar] [CrossRef] [PubMed]
- Mantzourani, M.; Fenlon, M.; Beighton, D. Association between Bifidobacteriaceae and the clinical severity of root caries lesions. Oral Microbiol. Immunol. 2009, 24, 32–37. [Google Scholar] [CrossRef] [PubMed]
- Lamont, R.J.; Koo, H.; Hajishengallis, G. The oral microbiota: Dynamic communities and host interactions. Nat. Rev. Microbiol. 2018, 16, 745–759. [Google Scholar] [CrossRef]
- Preza, D.; Olsen, I.; Aas, J.A.; Willumsen, T.; Grinde, B.; Paster, B.J. Bacterial profiles of root caries in elderly patients. J. Clin. Micobiol. 2008, 46, 2015–2021. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Preza, D.; Olsen, I.; Willumsen, T.; Boches, S.K.; Cotton, S.K.; Grinde, B.; Paster, B.J. Microarray analysis of the microflora of root caries in elderly. Eur. J. Clin. Microbiol. Infect. Dis. 2009, 28, 509–517. [Google Scholar] [CrossRef] [Green Version]
- Rôças, I.N.; Alves, F.R.F.; Rachid, F.R.F.; Rachid, C.T.C.C.; Lima, K.C.; Assunção, I.V.; Gomes, P.N.; Siqueira, J.F., Jr. Microbiome of deep dentinal caries lesions in teeth with symptomatic irreversible pulpitis. PLoS ONE 2016, 11, e0154653. [Google Scholar] [CrossRef]
- Wolff, D.; Frese, C.; Schoilew, K.; Dalpke, A.; Wolff, B.; Boutin, S. Amplicon-based microbiome study highlights the loss of diversity and the establishment of a set of species in patients with dentin caries. PLoS ONE 2019, 14, e0219714. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Obata, J.; Takeshita, T.; Shibata, Y.; Yamanaka, W.; Unemori, M.; Akamine, A.; Yamashita, Y. Identification of the microbiota in carious dentin lesions using 16S rRNA gene sequencing. PLoS ONE 2014, 9, e103712. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Qin, B.; Du, M.; Zhong, H.; Xu, Q.; Li, Y.; Zhang, P.; Fan, M. Extensive description and comparison of human supra-gingival microbiome in root caries and health. PLoS ONE 2015, 10, e0117064. [Google Scholar] [CrossRef] [Green Version]
- Caton, J.G.; Armitage, G.; Berglundh, T.; Chapple, I.L.C.; Jepsen, S.; Kornman, K.S.; Mealey, B.L.; Papapanou, P.N.; Sanz, M.; Tonetti, M.S. A new classification scheme for periodontal and peri-implant diseases and conditions—Introduction and key changes from the 1999 classification. J. Periodontol. 2018, 89 (Suppl. 1), S1–S8. [Google Scholar] [CrossRef] [PubMed]
- Beighton, D.; Lynch, E.; Heath, M.R. A microbiological study of primary root-caries lesions with different treatment needs. J. Dent. Res. 1993, 72, 623–629. [Google Scholar] [CrossRef]
- World Health Organization. Oral Health Surveys: Basic Methods, 5th ed.; World Health Organization: Geneva, Switzerland, 2013; p. 44. ISBN 9789241548649. [Google Scholar]
- Kianoush, N.; Adler, C.J.; Nguyen, K.T.; Browne, G.V.; Simonian, M.; Hunter, N. Bacterial profile of dentine caries and the impact of pH on bacterial population diversity. PLoS ONE 2014, 9, e92940. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kwon, T.G.; Lee, C.O.; Park, J.W.; Choi, S.Y.; Rijal, G.; Shin, H.I. Osteonecrosis associated with dental implants in patients undergoing bisphosphonate treatment. Clin. Oral Implant. Res. 2014, 25, 632–640. [Google Scholar] [CrossRef] [PubMed]
- Maki, T.; Ishikawa, A.; Mastunaga, T.; Pointing, S.B.; Saito, Y.; Kasai, T.; Watanabe, K.; Aoki, K.; Horiuchi, A.; Lee, K.C.; et al. Atmospheric aerosol deposition influences marine microbial communities in oligotrophic surface waters of the western Pacific Ocean. Deep-Sea Res. I 2016, 118, 37–45. [Google Scholar] [CrossRef] [Green Version]
- Hasegawa, T.; Takenaka, S.; Oda, M.; Domon, H.; Hiyoshi, T.; Sasagawa, K.; Ohsumi, T.; Hayashi, N.; Okamoto, Y.; Yamamoto, H.; et al. Sulfated vizantin causes detachment of biofilms composed mainly of the genus Streptococcus without affecting bacterial growth and viability. BMC Microbiol. 2020, 20, 361. [Google Scholar] [CrossRef] [PubMed]
- Hiranmayi, K.V.; Sirisha, K.; Ramoji Rao, M.V.; Sudhakar, P. Novel pathogens in periodontal microbiology. J. Pharm. Bioall. Sci. 2017, 9, 155–163. [Google Scholar] [CrossRef]
- Brailsford, S.R.; Shah, B.; Simons, D.; Gilbert, S.; Clark, D.; Ines, I.; Adams, S.E.; Allison, C.; Beighton, D. The predominant aciduric microflora of root-caries lesions. J. Dent. Res. 2001, 80, 1828–1833. [Google Scholar] [CrossRef]
- Aas, J.A.; Griffen, A.L.; Dardis, S.R.; Lee, A.M.; Olsen, I.; Dewhirst, F.E.; Leys, E.J.; Paster, B.J. Bacteria of dental caries in primary and permanent teeth in children and young adults. J. Clin. Microbiol. 2008, 46, 1407–1417. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuribayashi, M.; Kitasako, Y.; Matin, K.; Sadr, A.; Shida, K.; Tagami, J. Intraoral pH measurement of carious lesions with qPCR of cariogenic bacteria to differentiate caries activity. J. Dent. 2012, 40, 222–228. [Google Scholar] [CrossRef] [PubMed]
- Kianoush, N.; Nguyen, K.A.; Browne, G.V.; Simonian, M.; Hunter, N. pH gradient and distribution of streptococci, lactobacilli, prevotellae and fusobacteria in carious dentine. Clin. Oral Investig. 2014, 18, 659–669. [Google Scholar] [CrossRef] [Green Version]
- Kameda, M.; Abiko, Y.; Washio, J.; Tanner, A.C.R.; Kressirer, C.A.; Mizoguchi, I.; Takahashi, N. Sugar metabolism of Scardovia wiggsiae, a novel caries-associated bacterium. Front. Microbiol. 2020, 11, 479. [Google Scholar] [CrossRef] [PubMed]
- Damé-Teixeira, N.; Parolo, C.C.F.; Maltz, M.; Devine, D.; Do, T. Gene expression profile of Scardovia spp. in the metatranscriptome of root caries. Braz. Oral Res. 2020, 34, e042. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, N.; Yamada, T. Glucose and lactate metabolism by Actinomyces naeslundii. Crit. Rev. Oral Biol. Med. 1999, 10, 487–503. [Google Scholar] [CrossRef] [PubMed]
- Damé-Teixeira, N.; Parolo, C.C.F.; Maltz, M.; Tugnait, A.; Devine, D.; Do, T. Actinomyces spp. gene expression in root caries lesions. J. Oral Microbiol. 2016, 8, 32383. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Qi, Y.; Lo, E.C.; McGrath, C.; Mei, M.L.; Dai, R. Using next-generation sequencing to detect oral microbiome change following periodontal interventions: A systematic review. Oral Dis. 2020, 27, 1073–1089. [Google Scholar] [CrossRef] [PubMed]
- Solomon, S.M.; Timpu, D.; Forna, D.A.; Stefanache, M.A.M.; Martu, S.; Stoleriu, S. AFM comparative study of root surface morphology after three methods of scaling. Mater. Plast. 2016, 53, 546–549. [Google Scholar]
- Butera, A.; Gallo, S.; Maiorani, C.; Molino, D.; Chiesa, A.; Preda, C.; Esposito, F.; Scribante, A. Probiotic alternative to chlorhexidine in periodontal therapy; evaluation of clinical and microbiological parameters. Microorganisms 2020, 9, 69. [Google Scholar] [CrossRef] [PubMed]
- Seminario-Amez, M.; López-López, J.; Estrugo-Devesa, A.; Ayuso-Montero, R.; Jané-Salas, E. Probiotics and oral health: A systematic review. Med. Oral Patol. Oral Cir. Bucal 2017, 22, 3282–e288. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nicolae, V.; Neamiu, B.; Picu, O.; Stefanache, M.A.M.; Cioranu, V.S.I. The comparative evaluation of salivary biomarkers (Calcium, Phosphate, Salivary pH) in cario-resistance versus cario-activity. Rev. Chim. 2016, 67, 821–824. [Google Scholar]




| SG (n = 11) | GCB (n = 11) | p Value | |
|---|---|---|---|
| Age (mean ± SD) | 66.7 ± 5.2 | 69.0 ± 5.5 | 0.474 |
| Gender female (%) | 72.7 | 63.6 | 0.748 |
| Number of remaining teeth (mean ± SD) | 19.5 ± 5.7 | 18.9 ± 5.6 | 0.847 |
| Root caries teeth (mean ± SD) | 2.5 ± 2.2 | 2.7 ± 1.7 | 0.748 |
| O’ Leary’s plaque control record (%) | 27.43 ± 8.05 | 29.82 ± 8.21 | 0.401 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Takenaka, S.; Edanami, N.; Komatsu, Y.; Nagata, R.; Naksagoon, T.; Sotozono, M.; Ida, T.; Noiri, Y. Periodontal Pathogens Inhabit Root Caries Lesions Extending beyond the Gingival Margin: A Next-Generation Sequencing Analysis. Microorganisms 2021, 9, 2349. https://doi.org/10.3390/microorganisms9112349
Takenaka S, Edanami N, Komatsu Y, Nagata R, Naksagoon T, Sotozono M, Ida T, Noiri Y. Periodontal Pathogens Inhabit Root Caries Lesions Extending beyond the Gingival Margin: A Next-Generation Sequencing Analysis. Microorganisms. 2021; 9(11):2349. https://doi.org/10.3390/microorganisms9112349
Chicago/Turabian StyleTakenaka, Shoji, Naoki Edanami, Yasutaka Komatsu, Ryoko Nagata, Traithawit Naksagoon, Maki Sotozono, Takako Ida, and Yuichiro Noiri. 2021. "Periodontal Pathogens Inhabit Root Caries Lesions Extending beyond the Gingival Margin: A Next-Generation Sequencing Analysis" Microorganisms 9, no. 11: 2349. https://doi.org/10.3390/microorganisms9112349






