Gut Microbiota Parameters Potentially Useful in Clinical Perspective
Abstract
:1. The Search for Biomarkers
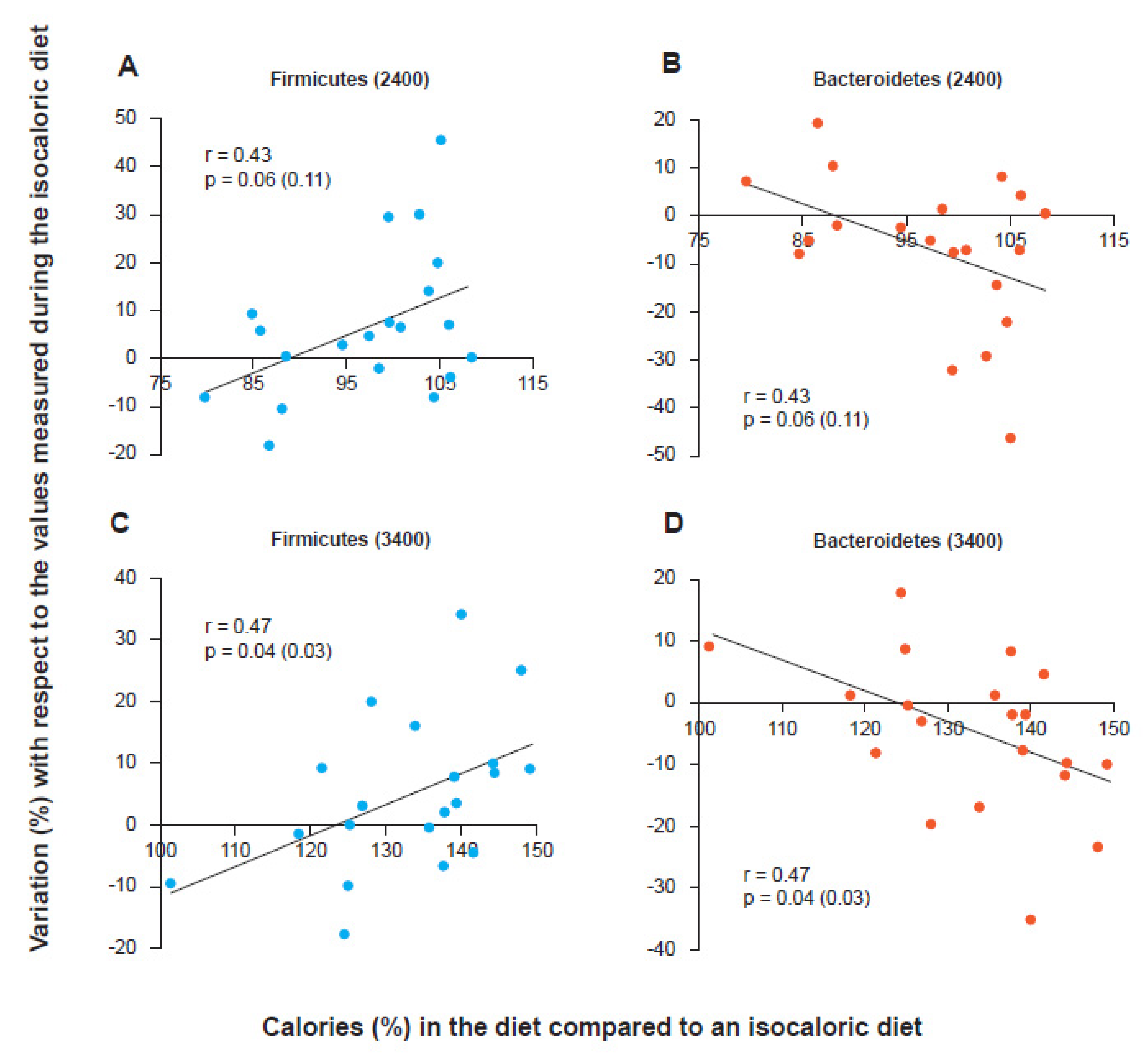
2. The Firmicutes/Bacteroidetes Ratio
3. Active Role of Firmicutes in the Predisposition to Weight Gain
4. Metabolic Endotoxemia
5. Gram-Positive and Gram-Negative
6. The Bacterial Lipopolysaccharide (LPS)
7. Toxicity and Immunogenicity of LPS
8. Role of the LPS Derived from Bacteroidetes
9. Gram-Negativity of the Colonic Microbiota and Metabolic Endotoxemia
10. The Gram-Positive/Gram-Negative Ratio
11. The Prevotella/Bacteroides Ratio
12. The Fusobacterium nucleatum/Faecalibacterium prausnitzii Ratio
13. Conclusions
Funding
Acknowledgments
Conflicts of Interest
References
- Haahtela, T.; Holgate, S.; Pawankar, R.; Akdis, C.A.; Benjaponpitak, S.; Caraballo, L.; Demain, J.; Portnoy, J.; von Hertzen, L.; WAO Special Committee on Climate Change and Biodiversity. The biodiversity hypothesis and allergic disease: World allergy organization position statement. World Allergy Organ. J. 2013, 6, 3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wilmanski, T.; Rappaport, N.; Earls, J.C.; Magis, A.T.; Manor, O.; Lovejoy, J.; Omenn, G.S.; Hood, L.; Gibbons, S.M.; Price, N.D. Blood metabolome predicts gut microbiome α-diversity in humans. Nat. Biotechnol. 2019, 37, 1217–1228. [Google Scholar] [CrossRef] [PubMed]
- Forslund, K.; Hildebrand, F.; Nielsen, T.; Falony, G.; Le Chatelier, E.; Sunagawa, S.; Prifti, E.; Vieira-Silva, S.; Gudmundsdottir, V.; Pedersen, H.K.; et al. Disentangling type 2 diabetes and metformin treatment signatures in the human gut microbiota. Nature 2015, 528, 262–266. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Li, Y.; Yan, L.; Sun, C.; Miao, Q.; Wang, Q.; Xiao, X.; Lian, M.; Li, B.; Chen, Y.; et al. Alterations of gut microbiome in autoimmune hepatitis. Gut 2020, 69, 569–577. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Li, F.; Liang, B.; Liang, Y.; Chen, S.; Mo, X.; Ju, Y.; Zhao, H.; Jia, H.; Spector, T.D.; et al. A metagenome-wide association study of gut microbiota in asthma in UK adults. BMC Microbiol. 2018, 18, 114. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, M.J.; Kang, M.J.; Lee, S.Y.; Lee, E.; Kim, K.; Won, S.; Suh, D.I.; Kim, K.W.; Sheen, Y.H.; Ahn, K.; et al. Perturbations of gut microbiome genes in infants with atopic dermatitis according to feeding type. J. Allergy Clin. Immunol. 2018, 141, 1310–1319. [Google Scholar] [CrossRef] [Green Version]
- Mancabelli, L.; Milani, C.; Lugli, G.A.; Turroni, F.; Cocconi, D.; van Sinderen, D.; Ventura, M. Identification of universal gut microbial biomarkers of common human intestinal diseases by meta-analysis. FEMS Microbiol. Ecol. 2017, 93, fix153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, Y.; Xu, Z.Z.; He, Y.; Yang, Y.; Liu, L.; Lin, Q.; Nie, Y.; Li, M.; Zhi, F.; Liu, S.; et al. Gut Microbiota Offers Universal Biomarkers across Ethnicity in Inflammatory Bowel Disease Diagnosis and Infliximab Response Prediction. Msystems 2018, 3, e00188-e17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, C.; Wang, H.; Liao, W.D.; Peng, C.; Shu, X.; Zhu, X.; Zhu, Z.H. Characteristics of mucosa-associated gut microbiota during treatment in Crohn’s disease. World J. Gastroenterol. 2019, 25, 2204–2216. [Google Scholar] [CrossRef] [PubMed]
- Louis, P.; Flint, H.J. Diversity, metabolism and microbial ecology of butyrate-producing bacteria from the human large intestine. FEMS Microbiol. Lett. 2009, 294, 1–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.; Li, Z.; He, Y.; Li, P.; Zhou, H.; Zeng, N. Regional distribution of Christensenellaceae and its associations with metabolic syndrome based on a population-level analysis. PeerJ 2020, 8, e9591. [Google Scholar] [CrossRef] [PubMed]
- Png, C.W.; Lindén, S.K.; Gilshenan, K.S.; Zoetendal, E.G.; McSweeney, C.S.; Sly, L.I.; McGuckin, M.A.; Florin, T.H. Mucolytic bacteria with increased prevalence in IBD mucosa augment in vitro utilization of mucin by other bacteria. Am. J. Gastroenterol. 2010, 105, 2420–2428. [Google Scholar] [CrossRef] [PubMed]
- Liang, J.Q.; Li, T.; Nakatsu, G.; Chen, Y.X.; Yau, T.O.; Chu, E.; Wong, S.; Szeto, C.H.; Ng, S.C.; Chan, F.K.L.; et al. A novel faecal Lachnoclostridium marker for the non-invasive diagnosis of colorectal adenoma and cancer. Gut 2020, 69, 1248–1257. [Google Scholar] [CrossRef] [Green Version]
- Alpizar-Rodriguez, D.; Lesker, T.R.; Gronow, A.; Gilbert, B.; Raemy, E.; Lamacchia, C.; Gabay, C.; Finckh, A.; Strowig, T. Prevotella copri in individuals at risk for rheumatoid arthritis. Ann. Rheum. Dis. 2019, 78, 590–593. [Google Scholar] [CrossRef] [PubMed]
- Mirza, A.; Forbes, J.D.; Zhu, F.; Bernstein, C.N.; Van Domselaar, G.; Graham, M.; Waubant, E.; Tremlett, H. The multiple sclerosis gut microbiota: A systematic review. Mult. Scler. Relat. Disord. 2020, 37, 101427. [Google Scholar] [CrossRef] [PubMed]
- Mirsepasi-Lauridsen, H.C.; Vallance, B.A.; Krogfelt, K.A.; Petersen, A.M. Escherichia coli Pathobionts Associated with Inflammatory Bowel Disease. Clin. Microbiol. Rev. 2019, 32, e00060-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roager, H.M.; Hansen, L.B.; Bahl, M.I.; Frandsen, H.L.; Carvalho, V.; Gøbel, R.J.; Dalgaard, M.D.; Plichta, D.R.; Sparholt, M.H.; Vestergaard, H.; et al. Colonic transit time is related to bacterial metabolism and mucosal turnover in the gut. Nat. Microbiol. 2016, 1, 16093. [Google Scholar] [CrossRef] [PubMed]
- Vandeputte, D.; Kathagen, G.; D’hoe, K.; Vieira-Silva, S.; Valles-Colomer, M.; Sabino, J.; Wang, J.; Tito, R.Y.; De Commer, L.; Darzi, Y.; et al. Quantitative microbiome profiling links gut community variation to microbial load. Nature 2017, 551, 507–511. [Google Scholar] [CrossRef]
- Valles-Colomer, M.; Falony, G.; Darzi, Y.; Tigchelaar, E.F.; Wang, J.; Tito, R.Y.; Schiweck, C.; Kurilshikov, A.; Joossens, M.; Wijmenga, C.; et al. The neuroactive potential of the human gut microbiota in quality of life and depression. Nat. Microbiol. 2019, 4, 623–632. [Google Scholar] [CrossRef] [PubMed]
- Allaband, C.; McDonald, D.; Vázquez-Baeza, Y.; Minich, J.J.; Tripathi, A.; Brenner, D.A.; Loomba, R.; Smarr, L.; Sandborn, W.J.; Schnabl, B.; et al. Microbiome 101: Studying, Analyzing, and Interpreting Gut Microbiome Data for Clinicians. Clin. Gastroenterol. Hepatol. 2019, 17, 218–230. [Google Scholar] [CrossRef] [PubMed]
- Ley, R.E.; Bäckhed, F.; Turnbaugh, P.; Lozupone, C.A.; Knight, R.D.; Gordon, J.I. Obesity alters gut microbial ecology. Proc. Natl. Acad. Sci. USA 2005, 102, 11070–11075. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turnbaugh, P.J.; Ley, R.E.; Mahowald, M.A.; Magrini, V.; Mardis, E.R.; Gordon, J.I. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 2006, 444, 1027–1031. [Google Scholar] [CrossRef] [PubMed]
- Ley, R.E.; Turnbaugh, P.J.; Klein, S.; Gordon, J.I. Microbial ecology: Human gut microbes associated with obesity. Nature 2006, 444, 1022–1023. [Google Scholar] [CrossRef]
- Bervoets, L.; Van Hoorenbeeck, K.; Kortleven, I.; Van Noten, C.; Hens, N.; Vael, C.; Goossens, H.; Desager, K.N.; Vankerckhoven, V. Differences in gut microbiota composition between obese and lean children: A cross-sectional study. Gut Pathog. 2013, 5, 10. [Google Scholar] [CrossRef] [Green Version]
- Koliada, A.; Syzenko, G.; Moseiko, V.; Budovska, L.; Puchkov, K.; Perederiy, V.; Gavalko, Y.; Dorofeyev, A.; Romanenko, M.; Tkach, S.; et al. Association between body mass index and Firmicutes/Bacteroidetes ratio in an adult Ukrainian population. BMC Microbiol. 2017, 17, 120. [Google Scholar] [CrossRef] [Green Version]
- Mariat, D.; Firmesse, O.; Levenez, F.; Guimarăes, V.; Sokol, H.; Doré, J.; Corthier, G.; Furet, J.P. The Firmicutes/Bacteroidetes ratio of the human microbiota changes with age. BMC Microbiol. 2009, 9, 123. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.S.; Hwang, S.S.; Park, E.J.; Bae, J.W. Strict vegetarian diet improves the risk factors associated with metabolic diseases by modulating gut microbiota and reducing intestinal inflammation. Environ. Microbiol. Rep. 2013, 5, 765–775. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Huang, Z.P.; Liu, C.Q.; Qi, L.; Sheng, Y.; Zou, D.J. Modulation of the gut microbiome: A systematic review of the effect of bariatric surgery. Eur. J. Endocrinol. 2018, 178, 43–56. [Google Scholar] [CrossRef] [PubMed]
- Magne, F.; Gotteland, M.; Gauthier, L.; Zazueta, A.; Pesoa, S.; Navarrete, P.; Balamurugan, R. The Firmicutes/Bacteroidetes Ratio: A Relevant Marker of Gut Dysbiosis in Obese Patients? Nutrients 2020, 12, 1474. [Google Scholar] [CrossRef] [PubMed]
- Jumpertz, R.; Le, D.S.; Turnbaugh, P.J.; Trinidad, C.; Bogardus, C.; Gordon, J.I.; Krakoff, J. Energy-balance studies reveal associations between gut microbes, caloric load, and nutrient absorption in humans. Am. J. Clin. Nutr. 2011, k94, 58–65. [Google Scholar] [CrossRef] [Green Version]
- Duncan, S.H.; Louis, P.; Thomson, J.M.; Flint, H.J. The role of pH in determining the species composition of the human colonic microbiota. Environ. Microbiol. 2009, 11, 2112–2122. [Google Scholar] [CrossRef]
- Bäckhed, F.; Ding, H.; Wang, T.; Hooper, L.V.; Koh, G.Y.; Nagy, A.; Semenkovich, C.F.; Gordon, J.I. The gut microbiota as an environmental factor that regulates fat storage. Proc. Natl. Acad. Sci. USA. 2004, 101, 15718–15723. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Flint, H.J.; Bayer, E.A.; Rincon, M.T.; Lamed, R.; White, B.A. Polysaccharide utilization by gut bacteria: Potential for new insights from genomic analysis. Nat. Rev. Microbiol. 2008, 6, 121–131. [Google Scholar] [CrossRef] [PubMed]
- Blaak, E.E.; Canfora, E.E.; Theis, S.; Frost, G.; Groen, A.K.; Mithieux, G.; Nauta, A.; Scott, K.; Stahl, B.; van Harsselaar, J.; et al. Short chain fatty acids in human gut and metabolic health. Benef. Microbes 2020, 11, 411–455. [Google Scholar] [CrossRef]
- Roediger, W.E. Role of anaerobic bacteria in the metabolic welfare of the colonic mucosa in man. Gut 1980, 21, 793–798. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chambers, E.S.; Morrison, D.J.; Frost, G. Control of appetite and energy intake by SCFA: What are the potential underlying mechanisms? Proc. Nutr. Soc. 2015, 74, 328–336. [Google Scholar] [CrossRef] [PubMed]
- Chambers, E.S.; Viardot, A.; Psichas, A.; Morrison, D.J.; Murphy, K.G.; Zac-Varghese, S.E.; MacDougall, K.; Preston, T.; Tedford, C.; Finlayson, G.S.; et al. Effects of targeted delivery of propionate to the human colon on appetite regulation, body weight maintenance and adiposity in overweight adults. Gut 2015, 64, 1744–1754. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vieira-Silva, S.; Falony, G.; Darzi, Y.; Lima-Mendez, G.; Garcia Yunta, R.; Okuda, S.; Vandeputte, D.; Valles-Colomer, M.; Hildebrand, F.; Chaffron, S.; et al. Species-function relationships shape ecological properties of the human gut microbiome. Nat. Microbiol. 2016, 1, 16088. [Google Scholar] [CrossRef]
- Cani, P.D.; Amar, J.; Iglesias, M.A.; Poggi, M.; Knauf, C.; Bastelica, D.; Neyrinck, A.M.; Fava, F.; Tuohy, K.M.; Chabo, C.; et al. Metabolic endotoxemia initiates obesity and insulin resistance. Diabetes 2007, 56, 1761–1772. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schütt, C. CD14. Int. J. Biochem. Cell Biol. 1999, 31, 545–549. [Google Scholar] [CrossRef]
- Borzęcka, K.; Płóciennikowska, A.; Björkelund, H.; Sobota, A.; Kwiatkowska, K. CD14 mediates binding of high doses of LPS but is dispensable for TNF-α production. Mediat. Inflamm. 2013, 2013, 824919. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goyal, A.K.; Bhat, M.; Sharma, M.; Garg, M.; Khairwa, A.; Garg, R. Effect of green tea mouth rinse on Streptococcus mutans in plaque and saliva in children: An in vivo study. J. Indian Soc. Pedod. Prev. Dent. 2017, 35, 41–46. [Google Scholar] [CrossRef] [PubMed]
- Vollmer, W.; Blanot, D.; de Pedro, M.A. Peptidoglycan structure and architecture. FEMS Microbiol. Rev. 2008, 32, 149–167. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ultee, E.; van der Aart, L.T.; Zhang, L.; van Dissel, D.; Diebolder, C.A.; van Wezel, G.P.; Claessen, D.; Briegel, A. Teichoic acids anchor distinct cell wall lamellae in an apically growing bacterium. Commun. Biol. 2020, 3, 314. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, N.; Sapra, A. Gram Staining; StatPearls Publishing: Treasure Island, FL, USA, 2021. [Google Scholar]
- Coico, R. Gram staining. Curr. Protoc. Microbiol. 2005. Appendix 3:Appendix 3C. [Google Scholar] [CrossRef]
- Megrian, D.; Taib, N.; Witwinowski, J.; Beloin, C.; Gribaldo, S. One or two membranes? Diderm Firmicutes challenge the Gram-positive/Gram-negative divide. Mol. Microbiol. 2020, 113, 659–671. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taib, N.; Megrian, D.; Witwinowski, J.; Adam, P.; Poppleton, D.; Borrel, G.; Beloin, C.; Gribaldo, S. Genome-wide analysis of the Firmicutes illuminates the diderm/monoderm transition. Nat. Ecol. Evol. 2020, 4, 1661–1672. [Google Scholar] [CrossRef] [PubMed]
- Raetz, C.R.; Whitfield, C. Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 2002, 71, 635–700. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oliveira, J.; Reygaert, W.C. Gram Negative Bacteria; StatPearls Publishing: Treasure Island, FL, USA, 2021. [Google Scholar]
- Rietschel, E.T.; Kirikae, T.; Schade, F.U.; Ulmer, A.J.; Holst, O.; Brade, H.; Schmidt, G.; Mamat, U.; Grimmecke, H.D.; Kusumoto, S.; et al. The chemical structure of bacterial endotoxin in relation to bioactivity. Immunobiology 1993, 187, 169–190. [Google Scholar] [CrossRef]
- Broeker, N.K.; Barbirz, S. Not a barrier but a key: How bacteriophages exploit host’s O-antigen as an essential receptor to initiate infection. Mol. Microbiol. 2017, 105, 353–357. [Google Scholar] [CrossRef] [Green Version]
- Lim, K.H.; Staudt, L.M. Toll-like receptor signaling. Cold Spring Harb. Perspect. Biol. 2013, 5, a011247. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ryu, J.K.; Kim, S.J.; Rah, S.H.; Kang, J.I.; Jung, H.E.; Lee, D.; Lee, H.K.; Lee, J.O.; Park, B.S.; Yoon, T.Y.; et al. Reconstruction of LPS Transfer Cascade Reveals Structural Determinants within LBP, CD14, and TLR4-MD2 for Efficient LPS Recognition and Transfer. Immunity 2017, 46, 38–50. [Google Scholar] [CrossRef] [Green Version]
- Ju, M.; Liu, B.; He, H.; Gu, Z.; Liu, Y.; Su, Y.; Zhu, D.; Cang, J.; Luo, Z. MicroRNA-27a alleviates LPS-induced acute lung injury in mice via inhibiting inflammation and apoptosis through modulating TLR4/MyD88/NF-κB pathway. Cell Cycle 2018, 17, 2001–2018. [Google Scholar] [CrossRef] [Green Version]
- Jandhyala, S.M.; Talukdar, R.; Subramanyam, C.; Vuyyuru, H.; Sasikala, M.; Nageshwar Reddy, D. Role of the normal gut microbiota. World J. Gastroenterol. 2015, 21, 8787–8803. [Google Scholar] [CrossRef]
- Delahooke, D.M.; Barclay, G.R.; Poxton, I.R. A re-appraisal of the biological activity of Bacteroides LPS. J. Med. Microbiol. 1995, 42, 102–112. [Google Scholar] [CrossRef] [PubMed]
- Antunes, L.C.; Poppleton, D.; Klingl, A.; Criscuolo, A.; Dupuy, B.; Brochier-Armanet, C.; Beloin, C.; Gribaldo, S. Phylogenomic analysis supports the ancestral presence of LPS-outer membranes in the Firmicutes. eLife 2016, 5, e14589. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Gao, Z.; Zhang, Z.; Pan, L.; Zhang, Y. Roles of M cells in infection and mucosal vaccines. Hum. Vaccin Immunother. 2014, 10, 3544–3551. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Akiba, Y.; Maruta, K.; Takajo, T.; Narimatsu, K.; Said, H.; Kato, I.; Kuwahara, A.; Kaunitz, J.D. Lipopolysaccharides transport during fat absorption in rodent small intestine. Am. J. Physiol. Gastrointest. Liver Physiol. 2020, 318, G1070–G1087. [Google Scholar] [CrossRef]
- Stevens, B.R.; Goel, R.; Seungbum, K.; Richards, E.M.; Holbert, R.C.; Pepine, C.J.; Raizada, M.K. Increased human intestinal barrier permeability plasma biomarkers zonulin and FABP2 correlated with plasma LPS and altered gut microbiome in anxiety or depression. Gut 2018, 67, 1555–1557. [Google Scholar] [CrossRef] [PubMed]
- Xie, G.; Wang, X.; Liu, P.; Wei, R.; Chen, W.; Rajani, C.; Hernandez, B.Y.; Alegado, R.; Dong, B.; Li, D.; et al. Distinctly altered gut microbiota in the progression of liver disease. Oncotarget 2016, 7, 19355–19366. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fernández-Musoles, R.; García Tejedor, A.; Laparra, J.M. Immunonutritional contribution of gut microbiota to fatty liver disease. Nutr. Hosp. 2020, 37, 193–206. [Google Scholar] [PubMed]
- Gomes, J.M.G.; Costa, J.A.; Alfenas, R.C.G. Metabolic endotoxemia and diabetes mellitus: A systematic review. Metabolism 2017, 68, 133–144. [Google Scholar] [CrossRef] [PubMed]
- Carnevale, R.; Sciarretta, S.; Valenti, V.; di Nonno, F.; Calvieri, C.; Nocella, C.; Frati, G.; Forte, M.; d’Amati, G.; Pignataro, M.G.; et al. Low-grade endotoxaemia enhances artery thrombus growth via Toll-like receptor 4: Implication for myocardial infarction. Eur. Heart J. 2020, 41, 3156–3165. [Google Scholar] [CrossRef]
- Kallio, K.A.; Hätönen, K.A.; Lehto, M.; Salomaa, V.; Männistö, S.; Pussinen, P.J. Endotoxemia, nutrition, and cardiometabolic disorders. Acta Diabetol. 2015, 52, 395–404. [Google Scholar] [CrossRef]
- Kasselman, L.J.; Vernice, N.A.; DeLeon, J.; Reiss, A.B. The gut microbiome and elevated cardiovascular risk in obesity and autoimmunity. Atherosclerosis 2018, 271, 203–213. [Google Scholar] [CrossRef]
- Maes, M.; Twisk, F.N.; Kubera, M.; Ringel, K.; Leunis, J.C.; Geffard, M. Increased IgA responses to the LPS of commensal bacteria is associated with inflammation and activation of cell-mediated immunity in chronic fatigue syndrome. J. Affect. Disord. 2012, 136, 909–917. [Google Scholar] [CrossRef] [PubMed]
- Storm-Larsen, C.; Stiksrud, B.; Eriksen, C.; Nowak, P.; Holm, K.; Thalme, A.; Dyrhol-Riise, A.M.; Brix, S.; Hov, J.R.; Trøseid, M. Microbial translocation revisited: Targeting the endotoxic potential of gut microbes in HIV-infected individuals. AIDS 2019, 33, 645–653. [Google Scholar] [CrossRef] [PubMed]
- Gomez-Llorente, M.A.; Romero, R.; Chueca, N.; Martinez-Cañavate, A.; Gomez-Llorente, C. Obesity and Asthma: A Missing Link. Int J. Mol. Sci. 2017, 18, 1490. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Penders, J.; Thijs, C.; Mommers, M.; Stobberingh, E.E.; Dompeling, E.; Reijmerink, N.E.; van den Brandt, P.A.; Kerkhof, M.; Koppelman, G.H.; Postma, D.S. Host-microbial interactions in childhood atopy: Toll-like receptor 4 (TLR4), CD14, and fecal Escherichia coli. J. Allergy Clin. Immunol. 2010, 125, 231–236.e5. [Google Scholar] [CrossRef] [PubMed]
- Sheen, Y.H.; Jee, H.M.; Kim, D.H.; Ha, E.K.; Jeong, I.J.; Lee, S.J.; Baek, H.S.; Lee, S.W.; Lee, K.J.; Lee, K.S. Serum zonulin is associated with presence and severity of atopic dermatitis in children, independent of total IgE and eosinophil. Clin. Exp. Allergy 2018, 48, 1059–1062. [Google Scholar] [CrossRef] [PubMed]
- Park, S.H.; Kim, K.A.; Ahn, Y.T.; Jeong, J.J.; Huh, C.S.; Kim, D.H. Comparative analysis of gut microbiota in elderly people of urbanized towns and longevity villages. BMC Microbiol. 2015, 15, 49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, K.A.; Jeong, J.J.; Yoo, S.Y.; Kim, D.H. Gut microbiota lipopolysaccharide accelerates inflamm-aging in mice. BMC Microbiol. 2016, 16, 9. [Google Scholar] [CrossRef] [Green Version]
- Lankelma, J.M.; van Vught, L.A.; Belzer, C.; Schultz, M.J.; van der Poll, T.; de Vos, W.M.; Wiersinga, W.J. Critically ill patients demonstrate large interpersonal variation in intestinal microbiota dysregulation: A pilot study. Intensive Care Med. 2017, 43, 59–68. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ricciuto, A.; Griffiths, A.M. Clinical value of fecal calprotectin. Crit Rev. Clin. Lab. Sci. 2019, 56, 307–320. [Google Scholar] [CrossRef] [PubMed]
- Caviglia, G.P.; Dughera, F.; Ribaldone, D.G.; Rosso, C.; Abate, M.L.; Pellicano, R.; Bresso, F.; Smedile, A.; Saracco, G.M.; Astegiano, M. Serum zonulin in patients with inflammatory bowel disease: A pilot study. Minerva Med. 2019, 110, 95–100. [Google Scholar] [CrossRef] [PubMed]
- Roager, H.M.; Licht, T.R.; Poulsen, S.K.; Larsen, T.M.; Bahl, M.I. Microbial enterotypes, inferred by the prevotella-to-bacteroides ratio, remained stable during a 6-month randomized controlled diet intervention with the new nordic diet. Appl. Environ. Microbiol. 2014, 80, 1142–1149. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hjorth, M.F.; Blædel, T.; Bendtsen, L.Q.; Lorenzen, J.K.; Holm, J.B.; Kiilerich, P.; Roager, H.M.; Kristiansen, K.; Larsen, L.H.; Astrup, A. Prevotella-to-Bacteroides ratio predicts body weight and fat loss success on 24-week diets varying in macronutrient composition and dietary fiber: Results from a post-hoc analysis. Int. J. Obes. 2019, 43, 149–157. [Google Scholar] [CrossRef] [Green Version]
- Christensen, L.; Roager, H.M.; Astrup, A.; Hjorth, M.F. Microbial enterotypes in personalized nutrition and obesity management. Am. J. Clin. Nutr. 2018, 108, 645–651. [Google Scholar] [CrossRef] [PubMed]
- Roberfroid, M.B. Caloric value of inulin and oligofructose. J. Nutr. 1999, 129 (Suppl. 7), 1436S–1437S. [Google Scholar] [CrossRef] [Green Version]
- De Filippo, C.; Di Paola, M.; Ramazzotti, M.; Albanese, D.; Pieraccini, G.; Banci, E.; Miglietta, F.; Cavalieri, D.; Lionetti, P. Diet, Environments, and Gut Microbiota. A Preliminary Investigation in Children Living in Rural and Urban Burkina Faso and Italy. Front. Microbiol. 2017, 8, 1979. [Google Scholar] [CrossRef] [Green Version]
- Chen, T.; Long, W.; Zhang, C.; Liu, S.; Zhao, L.; Hamaker, B.R. Fiber-utilizing capacity varies in Prevotella-versus Bacteroides-dominated gut microbiota. Sci. Rep. 2017, 7, 2594. [Google Scholar] [CrossRef] [PubMed]
- Brennan, C.A.; Garrett, W.S. Fusobacterium nucleatum: Symbiont, opportunist and oncobacterium. Nat. Rev. Microbiol. 2019, 17, 156–166. [Google Scholar] [CrossRef]
- Zagato, E.; Pozzi, C.; Bertocchi, A.; Schioppa, T.; Saccheri, F.; Guglietta, S.; Fosso, B.; Melocchi, L.; Nizzoli, G.; Troisi, J.; et al. Endogenous murine microbiota member Faecalibaculum rodentium and its human homologue protect from intestinal tumour growth. Nat. Microbiol. 2020, 5, 511–524. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Siles, M.; Duncan, S.H.; Garcia-Gil, L.J.; Martinez-Medina, M. Faecalibacterium prausnitzii: From microbiology to diagnostics and prognostics. ISME J. 2017, 11, 841–852. [Google Scholar] [CrossRef] [PubMed]
- Guo, S.; Li, L.; Xu, B.; Li, M.; Zeng, Q.; Xiao, H.; Xue, Y.; Wu, Y.; Wang, Y.; Liu, W.; et al. A Simple and Novel Fecal Biomarker for Colorectal Cancer: Ratio of Fusobacterium Nucleatum to Probiotics Populations, Based on Their Antagonistic Effect. Clin. Chem. 2018, 64, 1327–1337. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oliveira, A.A.; Farias, L.M.; Nicoli, J.R.; Costa, J.E.; Carvalho, M.A. Bacteriocin production by Fusobacterium isolates recovered from the oral cavity of human subjects with and without periodontal disease and of marmosets. Res. Microbiol. 1998, 149, 585–594. [Google Scholar] [CrossRef]
- Zhang, X.; Zhu, X.; Cao, Y.; Fang, J.Y.; Hong, J.; Chen, H. Fecal Fusobacterium nucleatum for the diagnosis of colorectal tumor: A systematic review and meta-analysis. Cancer Med. 2019, 8, 480–491. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Edgar, R.C. Accuracy of taxonomy prediction for 16S rRNA and fungal ITS sequences. PeerJ 2018, 6, e4652. [Google Scholar] [CrossRef]
- Edgar, R.C. Updating the 97% identity threshold for 16S ribosomal RNA OTUs. Bioinformatics 2018, 34, 2371–2375. [Google Scholar] [CrossRef]
- Callahan, B.J.; Wong, J.; Heiner, C.; Oh, S.; Theriot, C.M.; Gulati, A.S.; McGill, S.K.; Dougherty, M.K. High-throughput amplicon sequencing of the full-length 16S rRNA gene with single-nucleotide resolution. Nucleic Acids Res. 2019, 47, e103. [Google Scholar] [CrossRef] [Green Version]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quijada, N.M.; Rodríguez-Lázaro, D.; Eiros, J.M.; Hernández, M. TORMES: An automated pipeline for whole bacterial genome analysis. Bioinformatics 2019, 35, 4207–4212. [Google Scholar] [CrossRef] [PubMed]
- Balvočiūtė, M.; Huson, D.H. SILVA, RDP, Greengenes, NCBI and OTT—How do these taxonomies compare? BMC Genom. 2017, 18 (Suppl. 2), 114. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Edgar, R. Taxonomy annotation and guide tree errors in 16S rRNA databases. PeerJ 2018, 6, e5030. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roberfroid, M.; Gibson, G.R.; Delzenne, N. The biochemistry of oligofructose, a nondigestible fiber: An approach to calculate its caloric value. Nutr. Rev. 1993, 51, 137–146. [Google Scholar] [CrossRef]
- Jayashree, B.; Bibin, Y.S.; Prabhu, D.; Shanthirani, C.S.; Gokulakrishnan, K.; Lakshmi, B.S.; Mohan, V.; Balasubramanyam, M. Increased circulatory levels of lipopolysaccharide (LPS) and zonulin signify novel biomarkers of proinflammation in patients with type 2 diabetes. Mol. Cell Biochem. 2014, 388, 203–210. [Google Scholar] [CrossRef]
- Paoli, A.; Mancin, L.; Bianco, A.; Thomas, E.; Mota, J.F.; Piccini, F. Ketogenic Diet and Microbiota: Friends or Enemies? Genes 2019, 10, 534. [Google Scholar] [CrossRef] [Green Version]
- Carloni, S.; Bertocchi, A.; Mancinelli, S.; Bellini, M.; Erreni, M.; Borreca, A.; Braga, D.; Giugliano, S.; Mozzarelli, A.M.; Manganaro, D.; et al. Identification of a choroid plexus vascular barrier closing during intestinal inflammation. Science 2021, 374, 439–448. [Google Scholar] [CrossRef]
- Ayling, R.M.; Kok, K. Fecal Calprotectin. Adv. Clin. Chem. 2018, 87, 161–190. [Google Scholar]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Di Pierro, F. Gut Microbiota Parameters Potentially Useful in Clinical Perspective. Microorganisms 2021, 9, 2402. https://doi.org/10.3390/microorganisms9112402
Di Pierro F. Gut Microbiota Parameters Potentially Useful in Clinical Perspective. Microorganisms. 2021; 9(11):2402. https://doi.org/10.3390/microorganisms9112402
Chicago/Turabian StyleDi Pierro, Francesco. 2021. "Gut Microbiota Parameters Potentially Useful in Clinical Perspective" Microorganisms 9, no. 11: 2402. https://doi.org/10.3390/microorganisms9112402
APA StyleDi Pierro, F. (2021). Gut Microbiota Parameters Potentially Useful in Clinical Perspective. Microorganisms, 9(11), 2402. https://doi.org/10.3390/microorganisms9112402





