High Rates of Bacterial Pulmonary Co-Infections and Superinfections Identified by Multiplex PCR among Critically Ill COVID-19 Patients
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Design
2.2. Patient Selection
2.3. Microbiology Samples
2.4. Evaluation of FA Results as Positive
2.5. Statistical Analysis
3. Results
3.1. Patients
3.2. CAP and VAP Diagnosis
3.3. FA Results
3.4. Associated Outcomes Related to FA Results
3.5. Culture Results
3.6. Comparison between FA and Culture Results
3.7. Effect of FA Results on Antimicrobial Therapy
3.8. Effect of Post-FA Culture Result on Antimicrobial Therapy
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Grasselli, G.; Greco, M.; Zanella, A.; Albano, G.; Antonelli, M.; Bellani, G.; Bonanomi, E.; Cabrini, L.; Carlesso, E.; Castelli, G.; et al. Risk Factors Associated With Mortality Among Patients With COVID-19 in Intensive Care Units in Lombardy, Italy. JAMA Intern. Med. 2020, 180, 1345–1355. [Google Scholar] [CrossRef]
- Clancy, C.J.; Nguyen, M.H. Coronavirus Disease 2019, Superinfections, and Antimicrobial Development: What Can We Expect? Clin. Infect. Dis. 2020, 71, 2736–2743. [Google Scholar] [CrossRef]
- Gudiol, C.; Dura-Miralles, X.; Aguilar-Company, J.; Hernandez-Jimenez, P.; Martinez-Cutillas, M.; Fernandez-Aviles, F.; Machado, M.; Vazquez, L.; Martin-Davila, P.; de Castro, N.; et al. Co-infections and superinfections complicating COVID-19 in cancer patients: A multicentre, international study. J. Infect. 2021, 83, 306–313. [Google Scholar] [CrossRef]
- Barrasa, H.; Martin, A.; Maynar, J.; Rello, J.; Fernandez-Torres, M.; Aguirre-Quinonero, A.; Canut-Blasco, A.; Alava, C.-S.I. High rate of infections during ICU admission of patients with severe SARS-CoV-2 pneumonia: A matter of time? J. Infect. 2021, 82, 186–230. [Google Scholar] [CrossRef]
- Contou, D.; Claudinon, A.; Pajot, O.; Micaelo, M.; Longuet Flandre, P.; Dubert, M.; Cally, R.; Logre, E.; Fraisse, M.; Mentec, H.; et al. Bacterial and viral co-infections in patients with severe SARS-CoV-2 pneumonia admitted to a French ICU. Ann. Intensive Care 2020, 10, 119. [Google Scholar] [CrossRef] [PubMed]
- Kreitmann, L.; Monard, C.; Dauwalder, O.; Simon, M.; Argaud, L. Early bacterial co-infection in ARDS related to COVID-19. Intensive Care Med. 2020, 46, 1787–1789. [Google Scholar] [CrossRef] [PubMed]
- Cohen, R.; Finn, T.; Babushkin, F.; Geller, K.; Alexander, H.; Shapiro, M.; Uda, M.; Mostrchy, A.R.; Amash, R.; Shimoni, Z.; et al. High rate of bacterial respiratory tract co-infections upon admission amongst moderate to severe COVID-19 patients. Infect. Dis. 2021, in press. [Google Scholar] [CrossRef]
- Fattorini, L.; Creti, R.; Palma, C.; Pantosti, A.; Unit of Antibiotic, R.; Special, P.; Unit of Antibiotic, R.; Special Pathogens of the Department of Infectious Diseases. Bacterial coinfections in COVID-19: An underestimated adversary. Ann Ist Super Sanita 2020, 56, 359–364. [Google Scholar] [CrossRef]
- Morris, D.E.; Cleary, D.W.; Clarke, S.C. Secondary Bacterial Infections Associated with Influenza Pandemics. Front Microbiol. 2017, 8, 1041. [Google Scholar] [CrossRef] [Green Version]
- Elabbadi, A.; Turpin, M.; Gerotziafas, G.T.; Teulier, M.; Voiriot, G.; Fartoukh, M. Bacterial coinfection in critically ill COVID-19 patients with severe pneumonia. Infection 2021, 49, 559–562. [Google Scholar] [CrossRef] [PubMed]
- Hughes, S.; Troise, O.; Donaldson, H.; Mughal, N.; Moore, L.S.P. Bacterial and fungal coinfection among hospitalized patients with COVID-19: A retrospective cohort study in a UK secondary-care setting. Clin. Microbiol. Infect. 2020, 26, 1395–1399. [Google Scholar] [CrossRef]
- Langford, B.J.; So, M.; Raybardhan, S.; Leung, V.; Westwood, D.; MacFadden, D.R.; Soucy, J.R.; Daneman, N. Bacterial co-infection and secondary infection in patients with COVID-19: A living rapid review and meta-analysis. Clin. Microbiol. Infect. 2020, 26, 1622–1629. [Google Scholar] [CrossRef]
- Chen, X.; Zhao, B.; Qu, Y.; Chen, Y.; Xiong, J.; Feng, Y.; Men, D.; Huang, Q.; Liu, Y.; Yang, B.; et al. Detectable Serum Severe Acute Respiratory Syndrome Coronavirus 2 Viral Load (RNAemia) Is Closely Correlated With Drastically Elevated Interleukin 6 Level in Critically Ill Patients With Coronavirus Disease 2019. Clin. Infect. Dis. 2020, 71, 1937–1942. [Google Scholar] [CrossRef]
- Dong, X.; Cao, Y.Y.; Lu, X.X.; Zhang, J.J.; Du, H.; Yan, Y.Q.; Akdis, C.A.; Gao, Y.D. Eleven faces of coronavirus disease 2019. Allergy 2020, 75, 1699–1709. [Google Scholar] [CrossRef]
- Yang, X.; Yu, Y.; Xu, J.; Shu, H.; Xia, J.; Liu, H.; Wu, Y.; Zhang, L.; Yu, Z.; Fang, M.; et al. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: A single-centered, retrospective, observational study. Lancet Respir. Med. 2020, 8, 475–481. [Google Scholar] [CrossRef] [Green Version]
- Farfour, E.; Lecuru, M.; Dortet, L.; Le Guen, M.; Cerf, C.; Karnycheff, F.; Bonnin, R.A.; Vasse, M.; Lesprit, P. Carbapenemase-producing Enterobacterales outbreak: Another dark side of COVID-19. Am. J. Infect. Control 2020, 48, 1533–1536. [Google Scholar] [CrossRef]
- O’Toole, R.F. The interface between COVID-19 and bacterial healthcare-associated infections. Clin. Microbiol. Infect. 2021, in press. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, J.Y.; Le Pape, P.; Lopez, O.; Esquea, K.; Labiosa, A.L.; Alvarez-Moreno, C. Candida auris: A latent threat to critically ill patients with COVID-19. Clin. Infect. Dis. 2020. [Google Scholar] [CrossRef]
- Lai, C.C.; Wang, C.Y.; Hsueh, P.R. Co-infections among patients with COVID-19: The need for combination therapy with non-anti-SARS-CoV-2 agents? J. Microbiol. Immunol. Infect. 2020, 53, 505–512. [Google Scholar] [CrossRef] [PubMed]
- Clinical Microbiology Procedures Handbook, 4th ed.; ASM Press: Washington, DC, USA, 2016; pp. 297–316.
- The Clinical and Laboratory Standards Institute (Annapolis Junction, MD, USA 20701). Performance Standards for Antimicrobial Susceptibility Testing, 31st ed.; CLSI: Wayne, PA, USA, 2021. [Google Scholar]
- Lansbury, L.; Lim, B.; Baskaran, V.; Lim, W.S. Co-infections in people with COVID-19: A systematic review and meta-analysis. J. Infect. 2020, 81, 266–275. [Google Scholar] [CrossRef]
- Rawson, T.M.; Moore, L.S.P.; Zhu, N.; Ranganathan, N.; Skolimowska, K.; Gilchrist, M.; Satta, G.; Cooke, G.; Holmes, A. Bacterial and Fungal Coinfection in Individuals With Coronavirus: A Rapid Review To Support COVID-19 Antimicrobial Prescribing. Clin. Infect. Dis. 2020, 71, 2459–2468. [Google Scholar] [CrossRef]
- Feldman, C.; Anderson, R. The role of co-infections and secondary infections in patients with COVID-19. Pneumonia 2021, 13, 5. [Google Scholar] [CrossRef] [PubMed]
- De Santis, V.; Corona, A.; Vitale, D.; Nencini, C.; Potalivo, A.; Prete, A.; Zani, G.; Malfatto, A.; Tritapepe, L.; Taddei, S.; et al. Bacterial infections in critically ill patients with SARS-2-COVID-19 infection: Results of a prospective observational multicenter study. Infection 2021, in press. [Google Scholar] [CrossRef] [PubMed]
- Lim, Z.J.; Subramaniam, A.; Ponnapa Reddy, M.; Blecher, G.; Kadam, U.; Afroz, A.; Billah, B.; Ashwin, S.; Kubicki, M.; Bilotta, F.; et al. Case Fatality Rates for Patients with COVID-19 Requiring Invasive Mechanical Ventilation. A Meta-analysis. Am. J. Respir. Crit. Care Med. 2021, 203, 54–66. [Google Scholar] [CrossRef]
- Imam, Z.; Odish, F.; Gill, I.; O’Connor, D.; Armstrong, J.; Vanood, A.; Ibironke, O.; Hanna, A.; Ranski, A.; Halalau, A. Older age and comorbidity are independent mortality predictors in a large cohort of 1305 COVID-19 patients in Michigan, United States. J. Intern Med. 2020, 288, 469–476. [Google Scholar] [CrossRef] [PubMed]
- Gallo Marin, B.; Aghagoli, G.; Lavine, K.; Yang, L.; Siff, E.J.; Chiang, S.S.; Salazar-Mather, T.P.; Dumenco, L.; Savaria, M.C.; Aung, S.N.; et al. Predictors of COVID-19 severity: A literature review. Rev. Med. Virol. 2021, 31, 1–10. [Google Scholar] [CrossRef]
- Buehler, P.K.; Zinkernagel, A.S.; Hofmaenner, D.A.; Wendel Garcia, P.D.; Acevedo, C.T.; Gomez-Mejia, A.; Mairpady Shambat, S.; Andreoni, F.; Maibach, M.A.; Bartussek, J.; et al. Bacterial pulmonary superinfections are associated with longer duration of ventilation in critically ill COVID-19 patients. Cell Rep. Med. 2021, 2, 100229. [Google Scholar] [CrossRef] [PubMed]
- Maataoui, N.; Chemali, L.; Patrier, J.; Tran Dinh, A.; Le Fevre, L.; Lortat-Jacob, B.; Marzouk, M.; d’Humieres, C.; Rondinaud, E.; Ruppe, E.; et al. Impact of rapid multiplex PCR on management of antibiotic therapy in COVID-19-positive patients hospitalized in intensive care unit. Eur. J. Clin. Microbiol. Infect. Dis. 2021, 40, 2227–2234. [Google Scholar] [CrossRef]
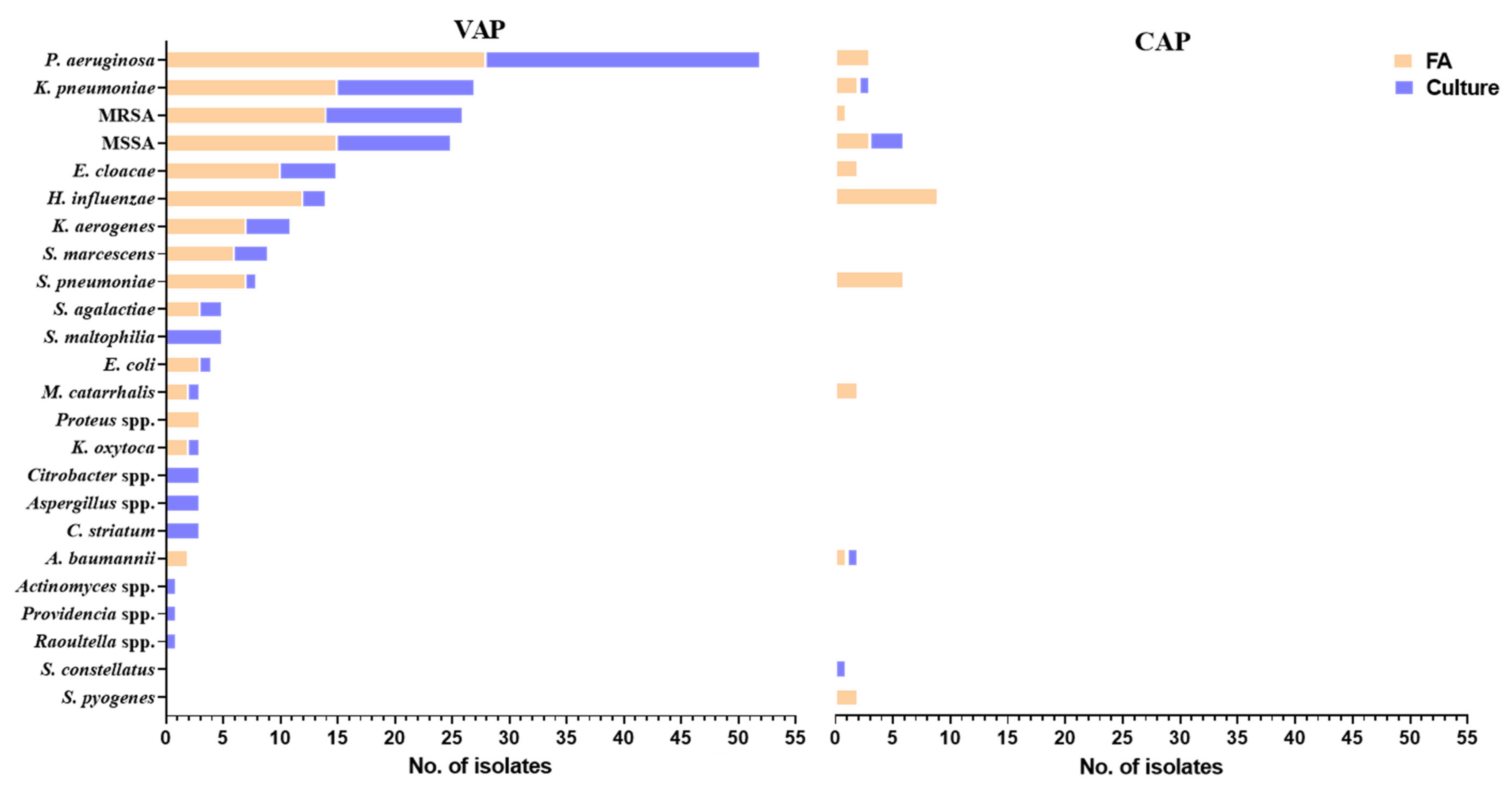
| Patients’ Characteristics | Number, Median (IQR) Total |
|---|---|
| Patients (n = 93) | |
| Age, years | 67 (56–76.5) |
| Male (%) | 65 (70) |
| LTCF residence, number (%) | 5 (5.3) |
| Length of stay, days | 24 (17–38) |
| COVID-19 diagnosis to hospitalization *, days (n = 87) | 3 (0–6) |
| Hospitalization to FA testing, days | 3 (1.5–6) |
| For diagnosis of CAP, days (n = 40) | 1 (1–2) |
| For diagnosis of VAP, days (n = 53) | 6 (4–9) |
| Patients diagnosed with ** (%): | |
| CAP | 16 (17) |
| VAP (without CAP) | 58 (62) |
| Neither | 19 (20) |
| C-reactive protein, mg/dL | 133 (76–220) |
| Mortality (%) | 36 (38.7) |
| Hospitalization to death, days (n = 36) | 25 (16–39.7) |
| VAP *** diagnosis to death, days (n = 33) | 17 (8.5–35.5) |
| Molecular and microbiology samples | |
| FA samples n = 148 (%) | |
| ETA | 102 (69) |
| Sputum | 22 (15) |
| BAL | 16 (11) |
| NBL | 8 (5) |
| Cultures samples n = 128 (%) | |
| ETA | 88 (69) |
| Sputum | 16 (12.5) |
| BAL | 16 (12.5) |
| NBL | 8 (6) |
| Patients with Only CAP n = 10 | Patients with VAP * n = 64 | Patients with Neither CAP nor VAP, n = 19 | CAP vs. VAP * p (95% CI ** or OR ***) | CAP vs. Neither p (95% CI or OR) | VAP * vs. Neither p (95% CI or OR) | |
|---|---|---|---|---|---|---|
| Age (years); median (IQR) | 57 (48.2–67.5) | 68.5 (63–77) | 59 (39–69) | 0.025 (1.1 to 17.5) | NS | 0.001 (−19.6 to −5) |
| Male (%) | 6 (60) | 48 (75) | 12 (63) | NS | NS | NS |
| LTCF residence (%) | 0 | 5 (7.8) | 0 | NS | NS | NS |
| Length of stay-days, median (IRQ) | 22.5 (16.2–29.5) | 29 (21–42.2) | 17 (12–29) | 0.08 (−1.3 to 23.3) | NS | 0.003 (−23.3 to −4.7) |
| Mortality ** (%) | 1 (10) | 33 (51.5) | 2 (10.5) | 0.001 (9.5) | NS | 0.0014 (9) |
| Strict Rules * | Less Strict Rules ** | |||||
|---|---|---|---|---|---|---|
| CAP (n = 12) | VAP (n = 62) | Total (n = 74) | CAP (n = 16) | VAP (n = 75) | Total (n = 91) | |
| Gram negative, number of targets detected (% ***) | ||||||
| E. cloacae | 2 (16.6) | 6 (9.6) | 8 (10.8) | 2 (12.5) | 10 (13.3) | 12 (13.1) |
| E. coli | 0 | 2 (3.2) | 2 (2.7) | 0 | 3 (4) | 3 (3.2) |
| A. baumannii | 1 (8.3) | 1 (1.6) | 2 (2.7) | 1 (6.2) | 2 (2.6) | 3 (3.2) |
| K. aerogenes | 0 | 5 (8) | 5 (6.7) | 0 | 7 (9.3) | 7 (7.6) |
| K. oxytoca | 0 | 0 | 0 | 0 | 2 (2.6) | 2 (2.1) |
| K. pneumoniae | 0 | 14 (22.5) | 14 (18.9) | 2 (12.5) | 15 (20) | 17 (18.6) |
| Proteus spp. | 0 | 2 (3.2) | 2 (2.7) | 0 | 3 (4) | 3 (3.2) |
| P. aeruginosa | 1 (8.3) | 26 (41.9) | 27 (36.4) | 3 (18.7) | 28 (37.3) | 31 (34) |
| M. catarrhalis | 2 (16.6) | 2 (3.2) | 4 (5.4) | 2 (12.5) | 2 (2.6) | 4 (4.3) |
| H. influenzae | 7 (58.3) | 8 (12.9) | 15 (20.2) | 9 (56.2) | 12 (16) | 21 (23) |
| S. marcescens | 0 | 4 (6.4) | 4 (5.4) | 0 | 6 (8) | 6 (6.5) |
| Total | 13 | 70 | 83 | 19 | 90 | 109 |
| Gram positive, number of targets detected (% ***) | ||||||
| S. agalactiae | 0 | 3 (4.8) | 3 (4) | 0 | 3 (4) | 3 (3.2) |
| S. pyogenes | 0 | 0 | 0 | 2 (12.5) | 0 | 2 (2.1) |
| S. aureus | 1 (8.3) | 23 (37) | 24 (32.4) | 4 (25) | 29 (38.6) | 33 (36.2) |
| MSSA | 1 | 11 | 12 | 3 | 15 | 18 |
| MRSA | 0 | 12 | 12 | 1 | 14 | 15 |
| S. pneumoniae | 6 (50) | 4 (6.4) | 10 (13.5) | 6 (37.5) | 7 (9.3) | 13 (14.2) |
| Total | 7 | 30 | 37 | 12 | 39 | 51 |
| Total | 20 | 100 | 120 | 31 | 129 | 160 |
| FA(+)/Culture(+) | FA(+)/Culture(−) | FA(−)/Culture(+) | FA(−)/Culture(−) | Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) | |
|---|---|---|---|---|---|---|---|---|
| Gram negative | ||||||||
| E. cloacae | 7/7 | 1/5 | 2/2 | 118/114 | 77.7/77.7 | 99.1/95.8 | 87.5/58.3 | 98.3/98.3 |
| E. coli | 1/1 | 0/1 | 0/0 | 127/126 | 100/100 | 100/99.2 | 100/50.0 | 100/100 |
| A. baumannii | 0/0 | 1/2 | 1/1 | 126/125 | 0/0 | 99.2/98.4 | 0/0 | 99.2/99.2 |
| K. aerogenes | 2/4 | 2/2 | 2/0 | 122/122 | 50/100 | 98.4/98.4 | 50/66.6 | 98.4/100 |
| K. oxytoca | 0/1 | 0/1 | 1/0 | 127/126 | 0/100 | 100/99.2 | -/50 | 99.2/100 |
| K. pneumoniae | 10/13 | 4/4 | 4/1 | 110/110 | 71.4/92.8 | 96.5/96.5 | 71.4/76.4 | 96.5/99.1 |
| Proteus spp. | 0/1 | 2/2 | 0/0 | 126/125 | -/100 | 98.4/98.4 | 0/33.3 | 100/100 |
| P. aeruginosa | 22/24 | 0/2 | 2/0 | 104/102 | 91.6/100 | 100/98 | 100/92.3 | 98.1/100 |
| M. catarrhalis | 1/1 | 1/1 | 0/0 | 126/126 | 100/100 | 99.2/99.2 | 50/50 | 100/100 |
| H. influenzae | 2/2 | 11/17 | 1/1 | 114/108 | 66.6/66.6 | 91.2/86.4 | 15.3/10.5 | 99.1/99.1 |
| S. marcescens | 1/2 | 2/3 | 0/0 | 125/123 | 100/100 | 98.4/97.6 | 33.3/40 | 100/100 |
| Total | 46/56 | 24/40 | 13/5 | 1325/1307 | 77.9/91.8 | 98.2/97 | 65.7/58.3 | 99.0/99.6 |
| Δ *** | +10 | +16 | −8 | −18 | +13.9 | −1.2 | −7.4 | +0.6 |
| Gram positive | ||||||||
| S. agalactiae | 2/2 | 1/1 | 0/0 | 125/125 | 100/100 | 99.2/99.2 | 66.6/66.6 | 100/100 |
| S. pyogenes | 0/1 | 0/1 | 1/0 | 127/126 | 0/100 | 100/99.2 | NA/50 | 99.2/100 |
| S. pneumoniae | 1/1 | 7/10 | 0/0 | 120/117 | 100/100 | 94.4/92.1 | 12.5/9.1 | 100/100 |
| S. aureus | 20/23 | 3/9 | 5/2 | 100/94 | 80/92 | 97.1/91.2 | 86.9/71.8 | 95.2/97.9 |
| MSSA | 9/11 | 2/6 | 4/2 | 113/109 | 69.2/84.6 | 98.3/94.8 | 81.8/64.7 | 96.6/98.2 |
| MRSA | 11/12 | 1/5 | 1/0 | 115/111 | 91.6/100 | 99.1/95.7 | 91.6/70.6 | 99.1/100 |
| Total | 23/27 | 11/21 | 6/2 | 472/462 | 79.3/93.1 | 97.7/95.6 | 67.6/56.2 | 98.7/99.5 |
| Δ | +4 | +10 | −4 | −10 | +13.8 | −2.1 | −11.4 | +0.8 |
| Total | 69/83 | 35/61 | 19/7 | 1797/1769 | 78.4/92.2 | 98.1/96.6 | 66.3/57.6 | 98.9/99.6 |
| Δ | +14 | +26 | −12 | −28 | +13.8 | −1.5 | −8.7 | +0.7 |
| Resistance genes | ||||||||
| CTX-M | 13/14 | 4/6 | 4/3 | 107/105 | 76.4/82.3 | 96.4/94.6 | 76.4/70 | 96.4/97.2 |
| NDM | 0/0 | 0/1 | 0/0 | 128/127 | - | 100/99.2 | NA/0 | 100/100 |
| IMP | 0/0 | 0/0 | 0/0 | 128/128 | - | 100/100 | - | 100/100 |
| KPC | 0/0 | 0/0 | 0/0 | 128/128 | - | 100/100 | - | 100/100 |
| OXA-48-like | 0/0 | 0/0 | 0/0 | 128/128 | - | 100/100 | - | 100/100 |
| Action Triggered by FA Result n (%) | Number of Patients | FA Results (n) | ABx Administered after FA Result | Corresponding Culture Results (n) | |||
|---|---|---|---|---|---|---|---|
| Avoiding ABx 21 (28.0) | 21 (100.0) | Negative FA | N/A | Normal flora (20) H. influenzae (1) | |||
| Starting ABx 41 (54.7) | Patients not treated previously 34 (82.9) | 18 (52.9) | H. influenzae (11) S. pneumoniae (6) MSSA (5) M. catarrhalis (2) | K. pneumoniae (2) E. coli (1) GBS (1) Negative FA (2) | ABx for community pathogens CRO (18) | MSSA (4) K. pnuemoniae (2) E. cloacae (2) S. pneumoniae (1) GBS (1) E. coli (1) | A. baumannii (1) H. influenzae (1) M. catarrhalis (1) Citrobacter spp. (1) Aspergillus spp. (1) Normal flora (8) |
| 7 (20.6) | P.aeruginosa (6) E. coli (1) E. cloacae (2) H. influenzae (1) | K. pneumoniae (2) MSSA (1) A. baumannii (1) Serratia spp. (1) | ABx for non-fermenters CAZ (5), TZP (2) | P.aeruginosa (3) E. cloacae (2) MSSA (1) | K. pnuemoniae (1) Serratia spp. (1) Normal flora (2) | ||
| 4 (11.8) | MSSA (3, one was sputum 104) H. influenzae (2) S. pneumoniae (1) GAS (1) Negative FA (1) | ABx for MSSA CFZ (3), AMC (1) | MSSA (2) Normal flora (1) | ||||
| 1 (2.9) | K. pneumoniae, CTX-M (1) | ABx for ESBL ETP (1) | K. pneumoniae (CRO-Res) | ||||
| 4 (11.8) | MRSA (4, one was sputum 105) P. aeruginosa (1) GAS (1) | ABx for MRSA VAN (4) | MRSA (2) P. aeruginosa (1) Aspergillus spp. (1) MSSA (1) Normal flora (1) | ||||
| Patients in “ABx window” 7 (17.1) | 1 (14.3) | MRSA, P.aeruginosa | VAN + TZP | MRSA, P.aeruginosa | |||
| 1 (14.3) | MRSA, E. cloacae (ETA—104), CTX-M | VAN | MRSA, S. maltophilia | ||||
| 1 (14.3) | MSSA, K. pneumoniae | CIP + CFZ | MSSA, K. pneumoniae (CRO-Sus) | ||||
| 1 (14.3) | K. aerogenes | TZP | K. aerogenes (CRO-Sus) | ||||
| 1 (14.3) | MRSA, E. cloacae, CTX-M | VAN + CIP | MRSA, E. cloacae (CRO-Res) | ||||
| 1 (14.3) | MRSA, K. aerogenes | VAN + MEM | K. aerogenes (CRO-Res) C. striatum | ||||
| 1 (14.3) | P. aeruginosa | CAZ | P. aeruginosa | ||||
| Stopping ABx 13 (17.3) | 11 (84.6) | Negative FA (11) | TZP (2) CXM + AZM (1) CRO (4) CRO + LVX (2) LVX (1) CHL (1) | Normal flora/yeasts (10) N/A (1) | |||
| 2 (15.4) | K. aerogenes (1) MRSA (1) | MEM (1) TZP (1) | K. aerogenes (1) MRSA (1) | ||||
| Action Triggered by FA Result n (%) | Number of Patients | FA Results (n) | ABx Given before FA Results | ABx Given after FA Results | Culture Results (n) | |
|---|---|---|---|---|---|---|
| Downgrading ABx 5 (6.8) | 1 | E. cloacae, P. aeruginosa | LVX | CIP | P. aeruginosa | |
| 1 | MSSA | TZP | OXA | Citrobacter spp. | ||
| 1 | Negative FA | VAN + TZP | TZP | Normal flora | ||
| 1 | MSSA | CRO | CFZ | MSSA | ||
| 1 | Negative FA | CRO + LVX | CRO | Normal flora | ||
| Upgrading ABx 27 (37.0) | For P. aeruginosa coverage 13 (48.1) | 13 | P. aeruginosa (13) and: K. pneumoniae (3) Serratia spp. (3) MSSA (2) MRSA (1) K. aerogenes (2) Proteus spp. (2) H. influenzae (2) E. cloacae (1) A. baumannii (1) CRO-M (2) | CRO (5) VAN & LVX (1) VAN (1) CRO + AZM (1) LVX (1) CAZ (1) CHL (2) ERT + VAN (1) CXM (1) | TZP (8) MEM (1) MEM + VAN (1) MEM + CST (1) CRO + LVX (1) CIP (1) | P. aeruginosa (10), MSSA (2), MRSA (1), K. pneumoniae CRO-Res (1), Providencia spp. (1), Roultella spp. (1), K. aerogenes (1), Normal flora (1), N/A (1) |
| For MRSA coverage 4 (14.8) | 1 | MRSA, K. oxytoca, CTX-M | CRO + AZM | SXT | MRSA, K. oxytoca CRO-Res | |
| 1 | MRSA, K. oxytoca, CTX-M | CRO | VAN + TZP | MRSA, E. cloacae CRO-Res | ||
| 1 | MRSA | TZP | VAN + LVX | MRSA | ||
| 1 | MRSA, K. pneumoniae | ERT | SXT | K. pneumoniae Non-CP-CRE | ||
| For ESBL coverage 4 (14.8) | 1 | CTX-M, E. cloacae, E. coli, Serratia spp. | SXT | MEM | Serratia spp., S. maltophilia | |
| 1 | CTX-M, E. cloacae | AZM | TZP | E. cloacae CRO-Res | ||
| 1 | CTX-M, K. pneumoniae | ERT | MEM + LZD | Yeast | ||
| 1 | CTX-M, K. pneumoniae | TZP | MEM | K. pneumoniae CRO-Res | ||
| Others 6 (22.2) | 1 | Negative FA | CST | AMB + VAN | N/A | |
| 1 | Negative FA | CFZ | SXT | Normal flora | ||
| 1 | E. cloacae (Sputum 104) | AZM | CIP | NA | ||
| 1 | MSSA | TZP | TZP + CFZ | Normal flora | ||
| 1 | H. influenzae | AMP | CRO | Normal flora | ||
| 1 | H. influenzae, S. pneumoniae, MSSA | AMK (due to bacteremia) | CRO | MSSA | ||
| No change in ABx 41 (56.2) | Negative FA (n = 17) | 17 | Negative FA | CRO (9) TZP (2) CRO + VAN (1) CRO + LVX (1) LVX (1) VAN (1) VAN + VRC (1) CHL (1) | Same | Normal flora (9), MSSA (2), S. maltophilia (1), Actinomyces spp. (1), C. striatum (1), Aspergillus flavus (1), P. aeruginosa (1), S. constellatus (1) |
| P. aeruginosa in FA (n = 8) | 8 | P. aeruginosa and: CTX-M (3) MRSA (1) MSSA (1) Proteus spp. (1) K. pneumoniae (1) E. cloacae (1) K. aerogenes (1) | TZP (2) CAZ (2) CAZ + CST (1) CST (1) VAN + TZP (1) MEM + VAN (1) | Same | P. aeruginosa (8) and K. pneumoniae CRO-Res (1), MRSA (1), Citrobacter spp. (1) | |
| CTX-M in FA (n = 3) | 3 | E. cloacae (1) K. pneumoniae (1) Serratia spp. (1) | ERT (2) LVX (1) | Same | S. maltophilia (1), Normal flora (1), K. pneumoniae CRO-Res (1) | |
| MRSA in FA (n = 2, one was sputum 104) | 2 | MRSA and S. pneumoniae (2) | MEM + VAN (2) | Same | Normal flora (2) | |
| MSSA in FA (n = 2) | 2 | MSSA (1) MSSA and GBS and S. pneumoniae (1) | LVX (1) CRO (1) | Same | S. maltophilia (1), Normal flora (1) | |
| Others (n = 9) | 9 | H. influenzae (3) K. pneumoniae (3) M. catarrhalis (2) GBS (1) S. pneumoniae (1) A. baumannii (1) E. cloacae (1) K. aerogenes (1) | CRO (8) ERT (1) | Same | K. pneumoniae (2), E. cloacae CRO-Res & K. pneumoniae (1), GBS (1), K. aerogenes (1), C. striatum (1) | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cohen, R.; Babushkin, F.; Finn, T.; Geller, K.; Alexander, H.; Datnow, C.; Uda, M.; Shapiro, M.; Paikin, S.; Lellouche, J. High Rates of Bacterial Pulmonary Co-Infections and Superinfections Identified by Multiplex PCR among Critically Ill COVID-19 Patients. Microorganisms 2021, 9, 2483. https://doi.org/10.3390/microorganisms9122483
Cohen R, Babushkin F, Finn T, Geller K, Alexander H, Datnow C, Uda M, Shapiro M, Paikin S, Lellouche J. High Rates of Bacterial Pulmonary Co-Infections and Superinfections Identified by Multiplex PCR among Critically Ill COVID-19 Patients. Microorganisms. 2021; 9(12):2483. https://doi.org/10.3390/microorganisms9122483
Chicago/Turabian StyleCohen, Regev, Frida Babushkin, Talya Finn, Keren Geller, Hanna Alexander, Candice Datnow, Martina Uda, Maurice Shapiro, Svetlana Paikin, and Jonathan Lellouche. 2021. "High Rates of Bacterial Pulmonary Co-Infections and Superinfections Identified by Multiplex PCR among Critically Ill COVID-19 Patients" Microorganisms 9, no. 12: 2483. https://doi.org/10.3390/microorganisms9122483
APA StyleCohen, R., Babushkin, F., Finn, T., Geller, K., Alexander, H., Datnow, C., Uda, M., Shapiro, M., Paikin, S., & Lellouche, J. (2021). High Rates of Bacterial Pulmonary Co-Infections and Superinfections Identified by Multiplex PCR among Critically Ill COVID-19 Patients. Microorganisms, 9(12), 2483. https://doi.org/10.3390/microorganisms9122483






