Effect of Nitrogen Addition on Soil Microbial Functional Gene Abundance and Community Diversity in Permafrost Peatland
Abstract
:1. Introduction
2. Materials and Methods
2.1. Site Description
2.2. Experimental Design and Sampling
2.3. Microbial Functional Gene Abundance and Community Diversity Analysis
2.4. Soil Chemical Analysis
2.5. Statistical Analyses
3. Results
3.1. Soil Chemical Properties
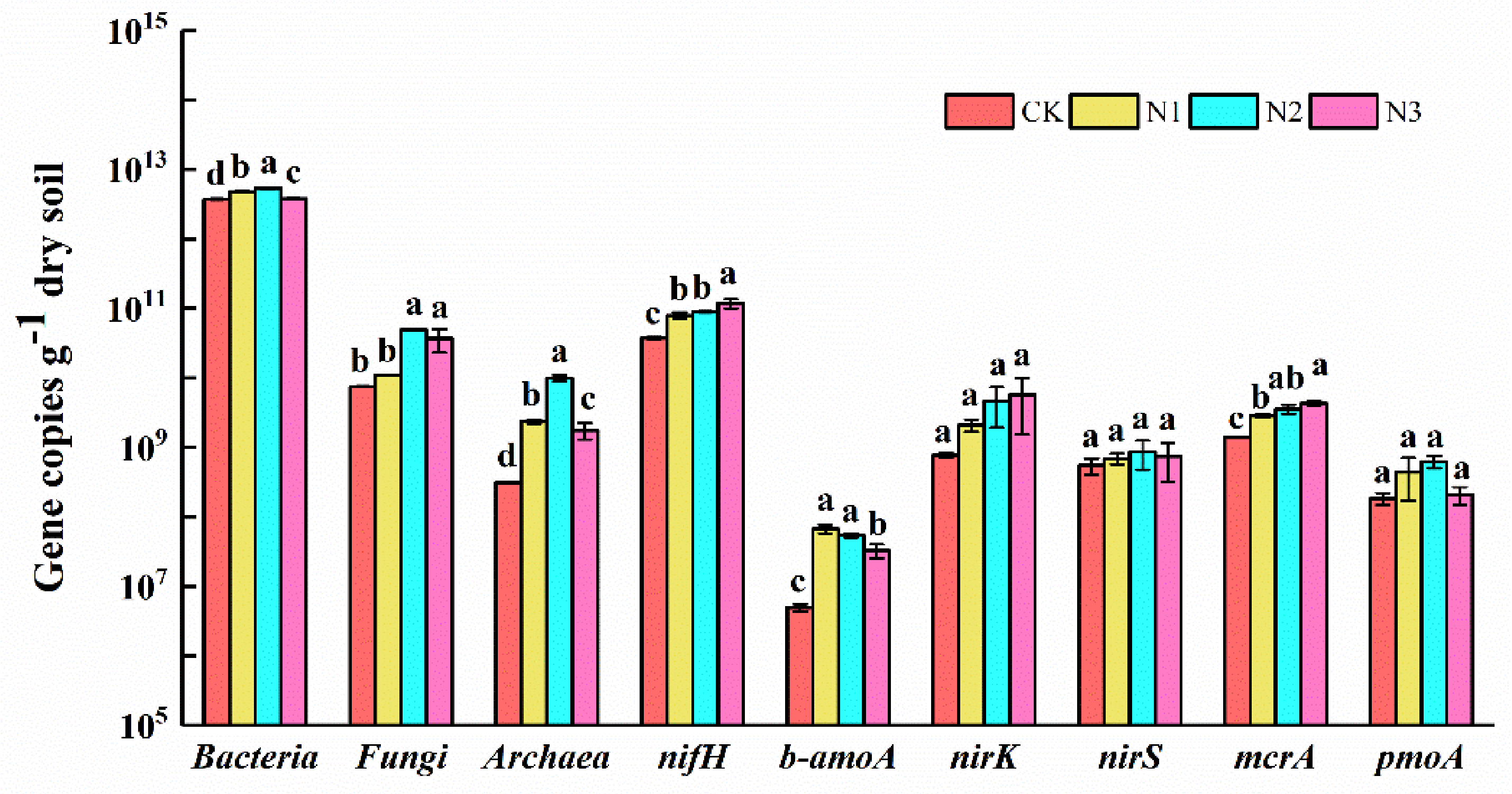
3.2. Soil Microbial Functional Group Abundance
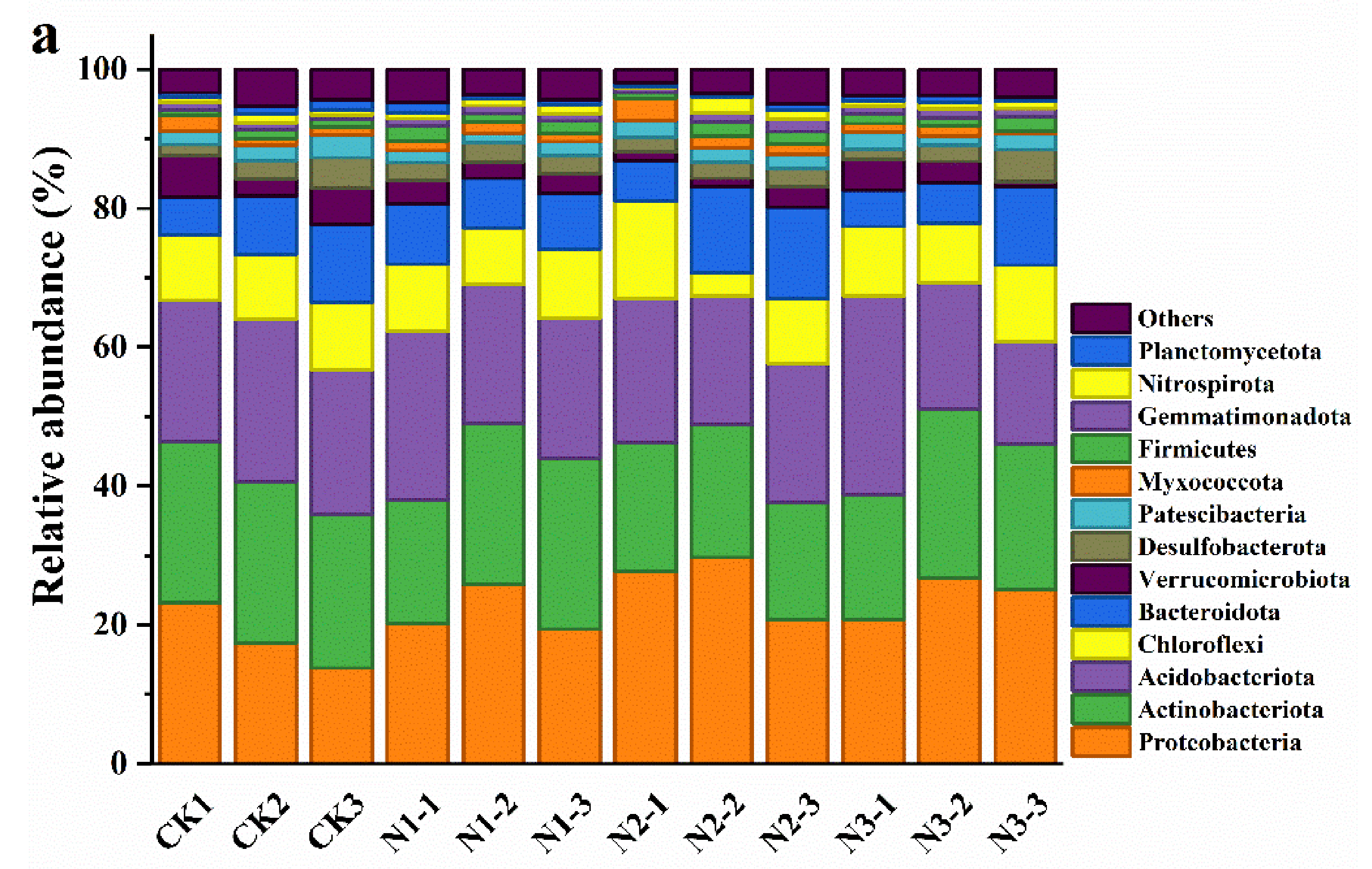
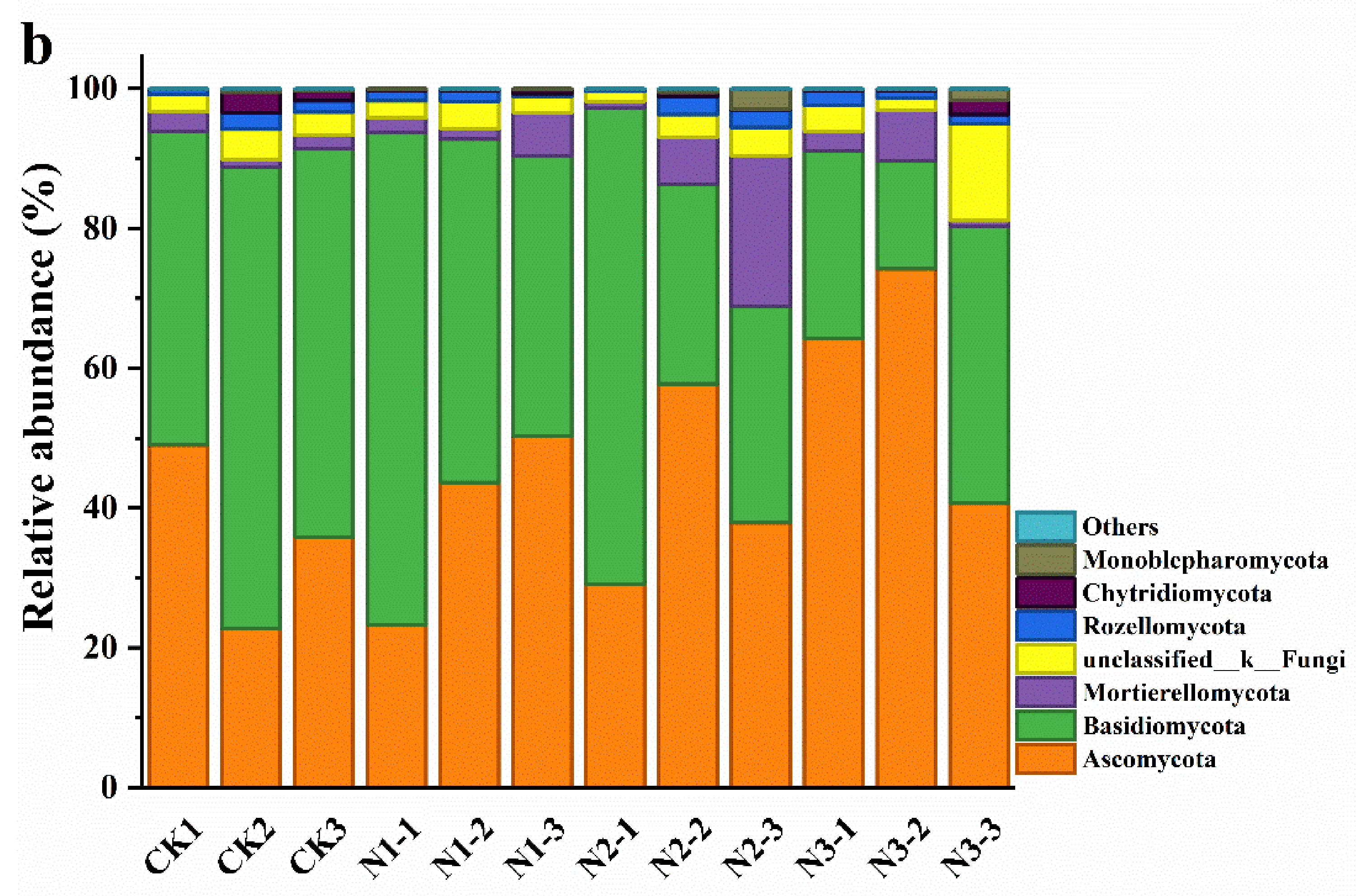
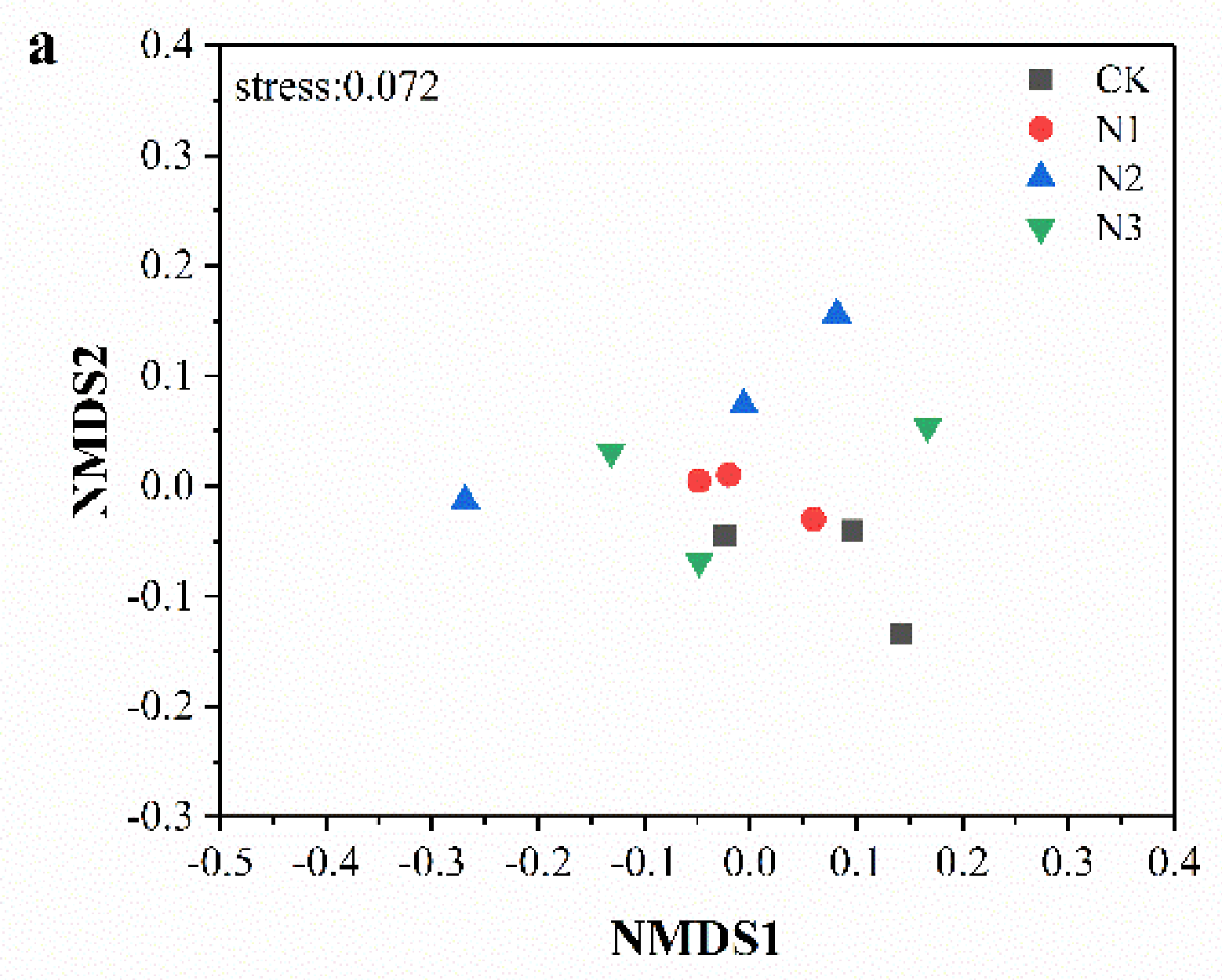
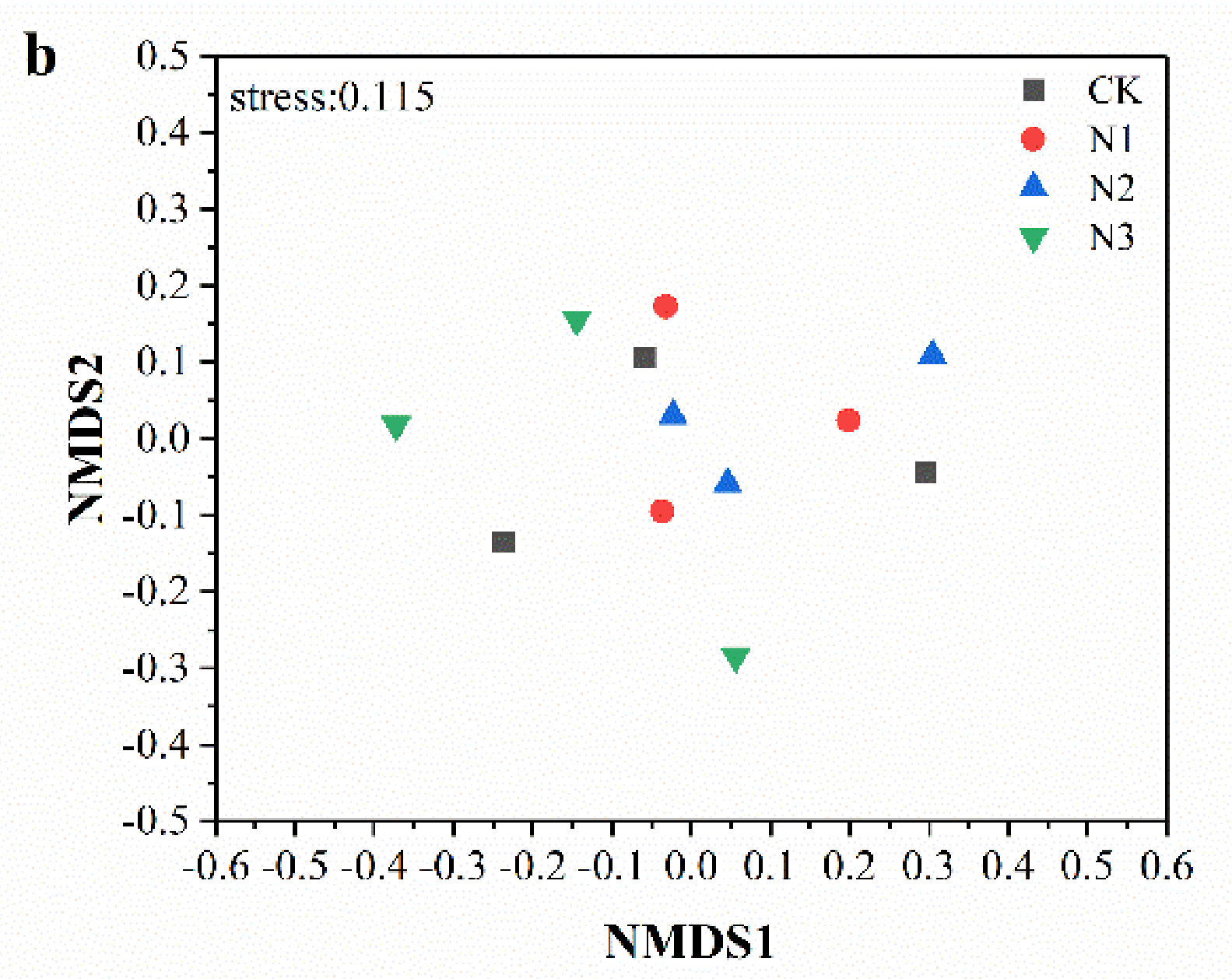
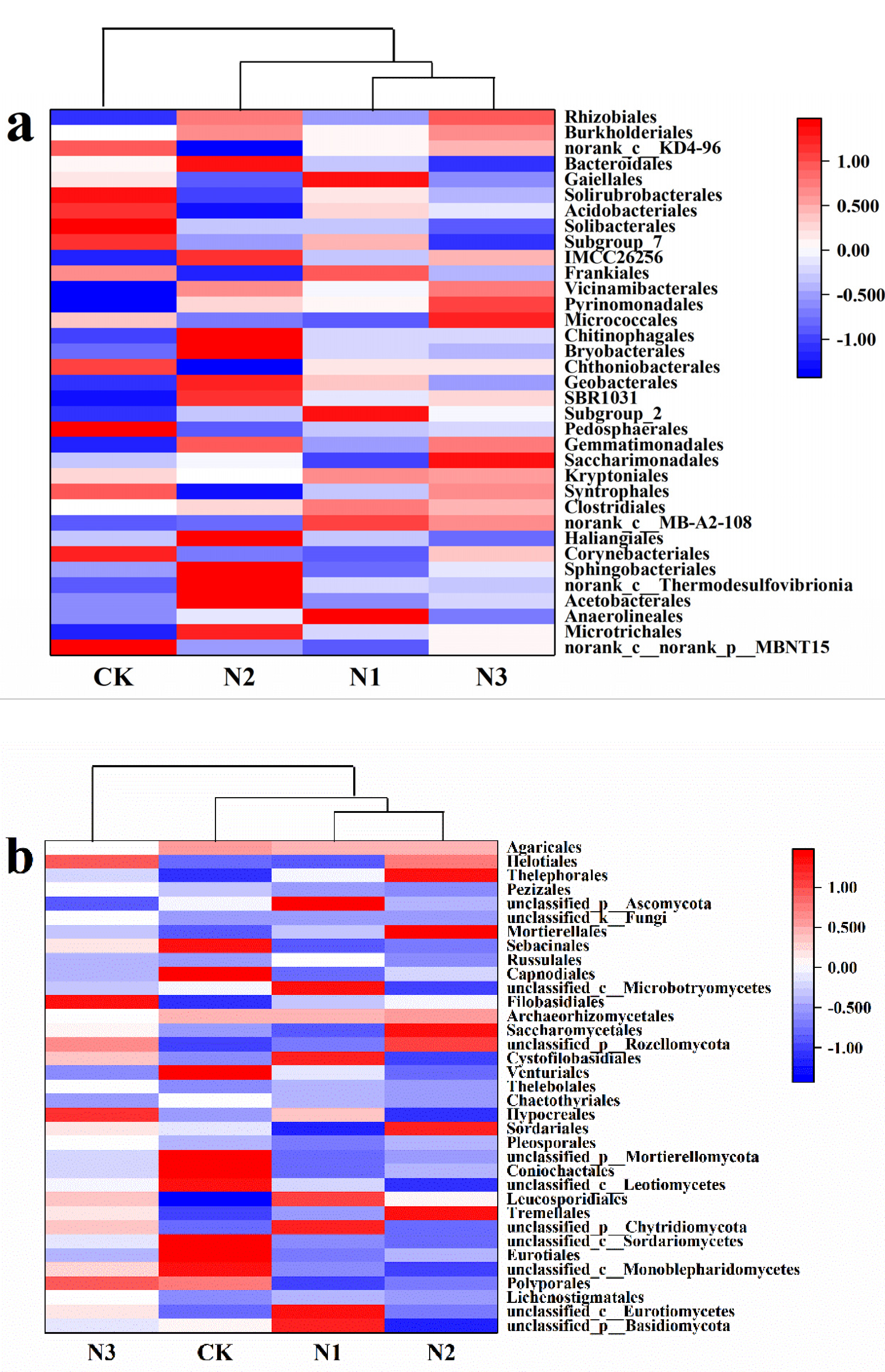
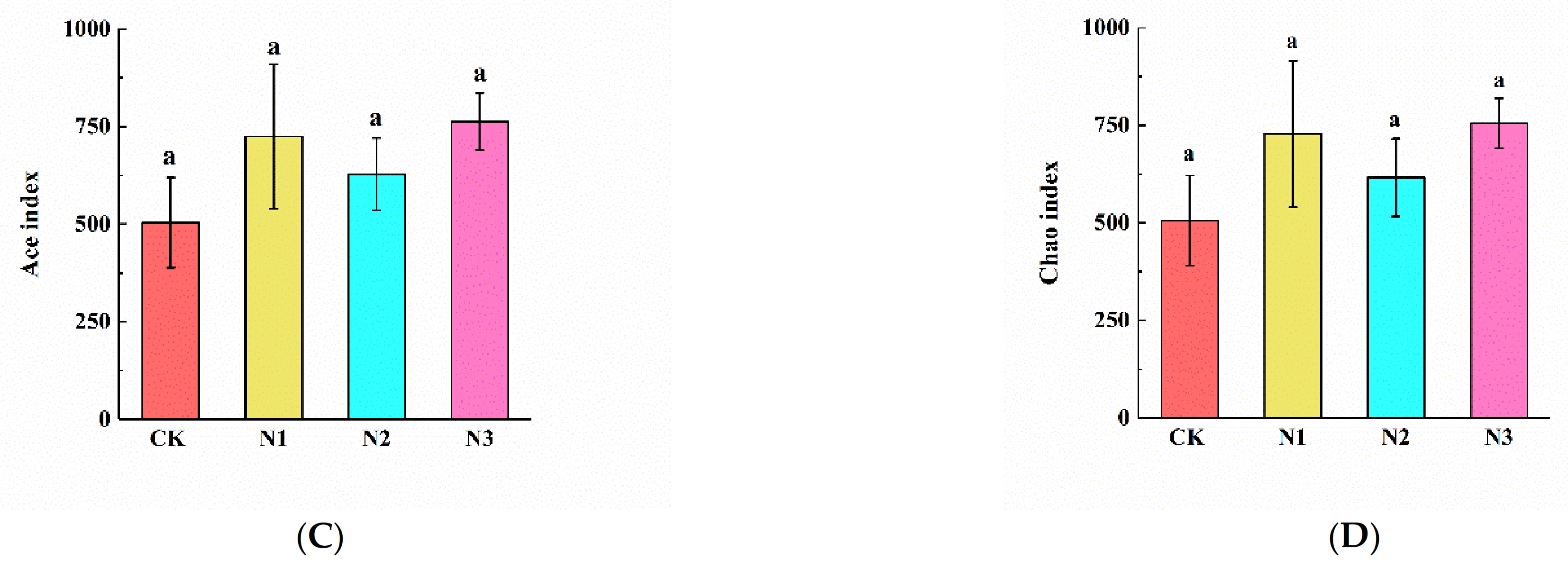
3.3. Soil Microbial Community Composition and Diversity
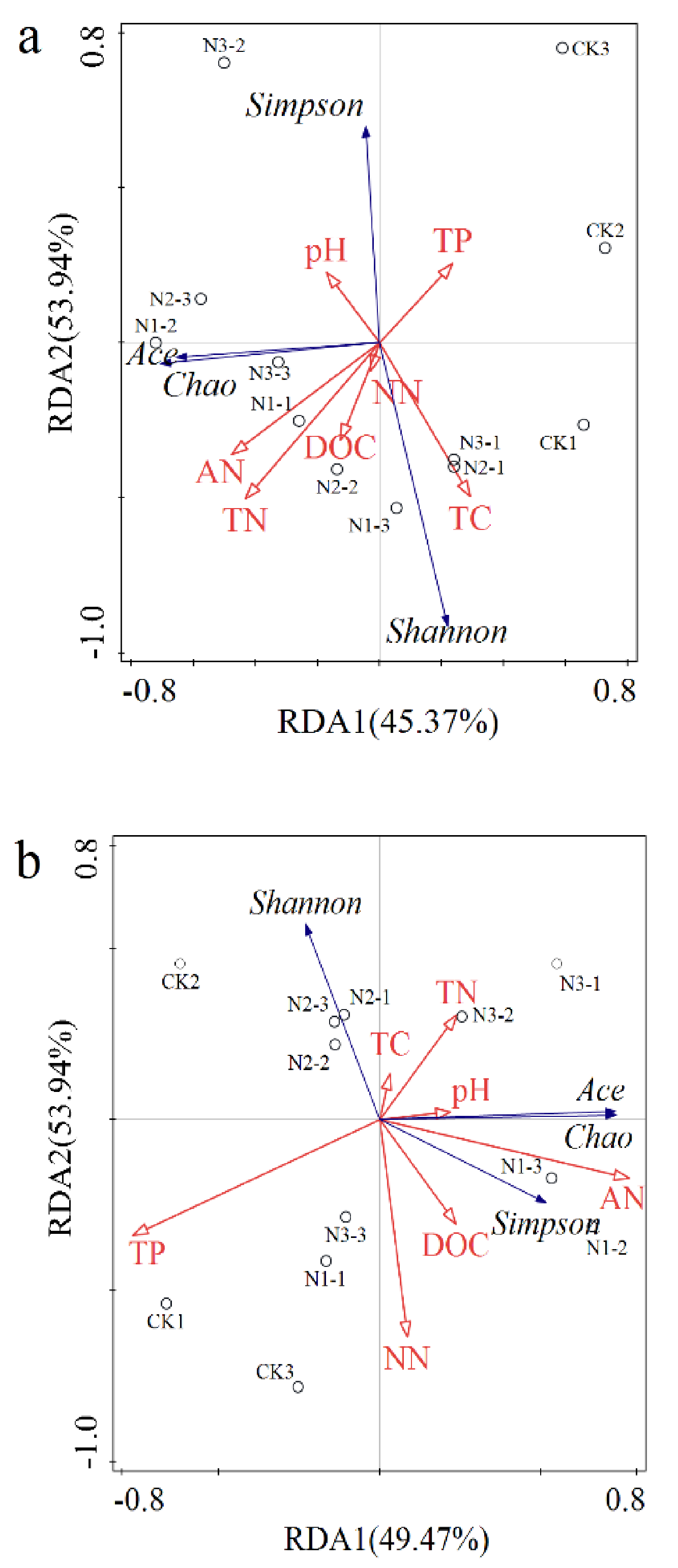
3.4. The Relationship between Soil Microorganisms and Soil Chemical Properties under Nitrogen Addition
| Indicator | Bacteria | Fungi | Archaea | nifH | B-amoA | nirK | nirS | mcrA | pmoA | TC | TN | TP | AN | NN | DOC | pH |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bacteria | 1 | |||||||||||||||
| Fungi | 0.42 | 1 | ||||||||||||||
| Archaea | 0.85 ** | 0.75 ** | 1 | |||||||||||||
| nifH | 0.17 | 0.68 * | 0.30 | 1 | ||||||||||||
| B-amoA | 0.75 ** | 0.31 | 0.51 | 0.46 | 1 | |||||||||||
| nirK | 0.14 | 0.73 ** | 0.36 | 0.73 ** | 0.27 | 1 | ||||||||||
| nirS | 0.31 | 0.48 | 0.35 | 0.44 | 0.28 | 0.78 ** | 1 | |||||||||
| mcrA | 0.26 | 0.73 ** | 0.41 | 0.94 ** | 0.48 | 0.74 ** | 0.47 | 1 | ||||||||
| pmoA | 0.81 ** | 0.42 | 0.65 * | 0.19 | 0.61 * | 0.13 | 0.21 | 0.22 | 1 | |||||||
| TC | −0.05 | −0.36 | −0.24 | −0.45 | 0.11 | −0.35 | −0.25 | −0.55 | 0.11 | 1 | ||||||
| TN | 0.86 ** | 0.75 ** | 0.92 ** | 0.36 | 0.63 * | 0.35 | 0.42 | 0.47 | 0.70 | −0.13 | 1 | |||||
| TP | 0.08 | −0.52 | −0.09 | −0.55 | −0.25 | −0.44 | −0.20 | −0.26 | 0.34 | 0.28 | −0.28 | 1 | ||||
| AN | −0.01 | −0.17 | −0.30 | 0.43 | 0.58 * | 0.10 | −0.02 | 0.36 | −0.04 | 0.22 | −0.09 | −0.32 | 1 | |||
| NN | 0.49 | −0.10 | 0.33 | −0.26 | 0.40 | −0.22 | 0.10 | −0.28 | 0.19 | 0.18 | 039 | 0.13 | 0.02 | 1 | ||
| DOC | 0.02 | −0.44 | −0.35 | −0.19 | 0.37 | −0.10 | 0.11 | −0.67 | 0.07 | 0.74 ** | 0.18 | 0.23 | 0.56 | 0.36 | 1 | |
| pH | −0.18 | 0.03 | −0.17 | 0.18 | 0.04 | −0.38 | −0.53 | 0.04 | 0.04 | 0.12 | −0.04 | −0.30 | 0.15 | −0.13 | −0.13 | 1 |

4. Discussion
4.1. Effect of N Addition on Soil Microbial Functional Gene Abundance in Permafrost Peatland
4.2. Effects of N Addition on Soil Microbial Community Composition and Diversity in Permafrost Peatland
4.3. Effects of Soil Chemical Properties on the Soil Microorganisms under Nitrogen Addition in Permafrost Peatland
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Elser, J.J.; Bracken, M.E.S.; Cleland, E.E.; Gruner, D.S.; Harpole, W.S.; Hillebrand, H.; Ngai, J.T.; Seabloom, E.W.; Shurin, J.B.; Smith, J.E. Global analysis of nitrogen and phosphorus limitation of primary producers in freshwater, marine and terrestrial ecosystems. Ecol. Lett. 2007, 10, 1135–1142. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, D.Z.; Zhao, B.; Wang, S.; Lei, D. Benefit of China’s reduction in nitrogen oxides emission to natural ecosystems in East Asia with respect to critical load exceedance. Environ. Int. 2020, 136, 105468. [Google Scholar] [CrossRef]
- Liu, X.J.; Zhang, Y.; Han, W.X.; Tang, A.H.; Shen, J.L.; Cui, Z.L.; Vitousek, P.; Erisman, J.W.; Goulding, K.; Christie, P.; et al. Enhanced nitrogen deposition over China. Nature 2013, 494, 459–462. [Google Scholar] [CrossRef]
- Galloway, J.N.; Townsend, A.R.; Erisman, J.W.; Bekunda, M.; Cai, Z.; Freney, J.R.; Martinelli, L.A.; Seitzinger, S.P.; Sutton, M.A. Transformation of the nitrogen cycle: Recent trends, questions, and potential solutions. Science 2008, 320, 889–892. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lamarque, J.F.; Kiehl, J.T.; Brasseur, G.P.; Butler, T.; Cameron-Smith, P.; Collins, W.D.; Collins, W.J.; Granier, C.; Hauglustaine, D.; Hess, P.G.; et al. Assessing future nitrogen deposition and carbon cycle feedback using a multimodel approach: Analysis of nitrogen deposition. J. Geophys. Res. Atmos. 2005, 110, D19303. [Google Scholar] [CrossRef] [Green Version]
- Contribution of Working Groups I, II, and III to the Fourth Assessment Report of the Intergovernmental Panel on Climate Change. In Proceedings of the IPCC Climate Change 2014: Synthesis Report, Geneva, Switzerland, 1 November 2014.
- Tian, H.Q.; Melillo, J.; Lu, C.Q.; Kicklighter, D.; Liu, M.L.; Ren, W.; Xu, X.F.; Chen, G.S.; Zhang, C.; Pan, S.F.; et al. China’s terrestrial carbon balance: Contributions from multiple global change factors. Glob. Biogeochem. Cycles 2011, 25, GB1007. [Google Scholar] [CrossRef]
- Xia, Z.; Yang, J.; Sang, C.; Wang, X.; Bai, E. Phosphorus reduces negative effects of nitrogen addition on soil microbial communities and functions. Microorganisms 2020, 8, 1828. [Google Scholar] [CrossRef]
- Craig, H.; Antwis, R.E.; Cordero, I.; Ashworth, D.; Robinson, C.H.; Osborne, T.Z.; Bardgett, R.D.; Rowntree, J.K.; Simpson, L.T. Nitrogen addition alters composition, diversity, and functioning of microbial communities in mangrove soils: An incubation experiment. Soil Biol. Biochem. 2021, 153, 108076. [Google Scholar] [CrossRef]
- Wang, C.; Liu, D.; Bai, E. Decreasing soil microbial diversity is associated with decreasing microbial biomass under nitrogen addition. Soil Biol. Biochem. 2018, 120, 126–133. [Google Scholar] [CrossRef]
- Bassiri, R.H. Consequences of atmospheric nitrogen deposition in terrestrial ecosystems, old questions, new perspectives. Oecologia 2015, 177, 1–3. [Google Scholar] [CrossRef] [Green Version]
- Damashek, J.; Francis, C.A. Microbial nitrogen cycling in Estuaries: From genes to ecosystem processes. Estuaries Coasts 2018, 41, 626–660. [Google Scholar] [CrossRef]
- Huang, L.; Riggins, C.W.; Rodríguez-Zas, S.; Zabaloy, M.C.; Villamil, M.B. Long-term N fertilization imbalances potential N acquisition and transformations by soil microbes. Sci. Total Environ. 2019, 691, 562–571. [Google Scholar] [CrossRef]
- Li, H.; Xu, Z.; Yang, S.; Li, X.; Top, E.M.; Wang, R.; Zhang, Y.; Cai, J.; Yao, F.; Han, X.; et al. Response of soil bacterial communities to nitrogen deposition and precipitation increment are closely linked with aboveground communtiy variation. Microb. Ecol. 2016, 71, 974–989. [Google Scholar] [CrossRef]
- Zhang, X.L.; Zhang, Q.Q.; Yang, A.J.; Hou, L.J.; Zheng, Y.L.; Zhai, W.D.; Gong, J. Incorporation of microbial functional traits in biogeochemistry models provides better estimations of benthic denitrification and anammox rates in coastal oceans. Biogeosciences 2018, 12, 3331–3352. [Google Scholar] [CrossRef]
- Fierer, N.; Lauber, C.L.; Ramirez, K.S.; Zaneveld, J.; Bradford, M.A.; Knight, R. Comparative metagenomic, phylogenetic and physiological analyses of soil microbial communities across nitrogen gradients. ISME J. 2012, 6, 1007–1017. [Google Scholar] [CrossRef] [Green Version]
- Zheng, S.; Bian, H.; Quan, Q.; Xu, L.; Chen, Z.; He, N. Effect of nitrogen and acid deposition on soil respiration in a temperate forest in China. Geoderma 2018, 329, 82–90. [Google Scholar] [CrossRef]
- Wang, J.Q.; Shi, X.Z.; Zheng, C.Y.; Suter, H.; Huang, Z.Q. Different responses of soil bacterial and fungal communities to nitrogen deposition in a subtropical forest. Sci. Total Environ. 2021, 755, 142449. [Google Scholar] [CrossRef]
- Zhang, J.Y.; Ai, Z.M.; Liang, C.T.; Wang, G.L.; Xue, S. Response of soil microbial communities and nitrogen thresholds of Bothriochloa ischaemum to short-term nitrogen addition on the Loess Plateau. Geoderma 2017, 308, 112–119. [Google Scholar] [CrossRef]
- van der Bom, F.; Nunes, I.; Raymond, N.S.; Hansen, V.; Bonnichsen, L.; Magid, J.; Nybroe, O.; Jensen, L.S. Long-term fertilisation form, level and duration affect the diversity, structure and functioning of soil microbial communities in the field. Soil Biol. Biochem. 2018, 122, 91–103. [Google Scholar] [CrossRef]
- Ouyang, Y.; Norton, J.M. Short-term nitrogen fertilization affects microbial community composition and nitrogen mineralization functions in an agricultural soil. Appl. Environ. Microbiol. 2020, 86, e02278–e02319. [Google Scholar] [CrossRef]
- Tang, H.M.; Li, C.; Xiao, X.P.; Shi, L.H.; Cheng, K.K.; Wen, L.; Li, W.W. Effects of short-term manure nitrogen input on soil microbial community structure and diversity in a double-cropping paddy field of southern China. Sci. Rep. 2020, 10, 13540. [Google Scholar] [CrossRef] [PubMed]
- Li, J.H.; Cheng, B.H.; Zhang, R.; Li, W.J.; Shi, X.M.; Wei, H.Y.; Ye, L.F.; Ostle, N.J.; Bardgett, R.D. Nitrogen and phosphorus additions accelerate decomposition of slow carbon pool and lower total soil organic carbon pool in alpine meadows. Land Degrad. Dev. 2021, 32, 1761–1772. [Google Scholar] [CrossRef]
- Shi, X.Z.; Wang, J.Q.; Müller, C.; Hu, H.W.; He, J.Z.; Wang, J.T.; Huang, Z.Q. Dissimilatory nitrate reduction to ammonium dominates soil nitrate retention capacity in subtropical forests. Biol. Fertil. Soils 2020, 56, 785–797. [Google Scholar] [CrossRef]
- Moore, T.R.; Bubier, J.L. Plant and soil Nitrogen in an ombrotrophic peatland, Southern Canada. Ecosystems 2020, 23, 98–110. [Google Scholar] [CrossRef]
- Salmon, V.G.; Brice, D.J.; Bridgham, S.; Childs, J.; Graham, J.; Griffiths, N.A.; Hofmockel, K.; Iversen, C.M.; Jicha, T.M.; Kolka, R.K.; et al. Nitrogen and phosphorus cycling in an ombrotrophic peatland: A benchmark for assessing change. Plant Soil 2021, 466, 649–674. [Google Scholar] [CrossRef]
- Waldrop, M.P.; Zak, D.R.; Sinsabaugh, R.L.; Gallo, M.; Lauber, C. Nitrogen deposition modifies soil carbon storage through changes in microbial enzymatic activity. Ecol. Appl. 2004, 14, 1172–1177. [Google Scholar] [CrossRef]
- Mack, M.C.; Schuur, E.A.G.; Bret-Harte, M.S.; Shaver, G.R.; Chapin, F.S. Ecosystem carbon storage in arctic tundra reduced by long-term nutrient fertilization. Nature 2004, 431, 440–443. [Google Scholar] [CrossRef]
- Gong, Y.; Wu, J.H.; Vogt, J.; Le, T.B. Warming reduces the increase in N2O emission under nitrogen fertilization in a boreal peatland. Sci. Total Environ. 2019, 664, 72–78. [Google Scholar] [CrossRef]
- Miao, Y.; Song, C.; Sun, L.; Wang, X.; Meng, H.; Mao, R. Growing season methane emission from a boreal peatland in the continuous permafrost zone of Northeast China: Effects of active layer depth and vegetation. Biogeosciences 2012, 9, 4455–4464. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Z.; Schwartz, S.; Wagner, L.; Miller, W. A greedy algorithm for aligning DNA sequences. J. Comput. Biol. 2000, 7, 203–214. [Google Scholar] [CrossRef]
- Xu, N.; Tan, G.; Wang, H.; Gai, X. Effect of biochar additions to soil on nitrogen leaching, microbial biomass and bacterial community structure. Eur. J. Soil Biol. 2016, 74, 1–8. [Google Scholar] [CrossRef]
- Adams, R.I.; Miletto, M.; Taylor, J.W.; Bruns, T.D. Dispersal in microbes: Fungi in indoor air are dominated by outdoor air and show dispersal limitation at short distances. ISME J. 2013, 7, 1262–1273. [Google Scholar] [CrossRef] [Green Version]
- Magoč, T.; Salzberg, S. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [Green Version]
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahé, F. Vsearch: A versatile open source tool for metagenomics. Peer J. 2016, 4, e2584. [Google Scholar] [CrossRef]
- Edgar, R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef] [Green Version]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naive bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef] [Green Version]
- Ghani, A.; Dexter, M.; Perrott, K.W. Hot-water extractable carbon in soils: A sensitive measurement for determining impacts of fertilization, grazing and cultivation. Soil Biol. Biochem. 2003, 35, 1231–1243. [Google Scholar] [CrossRef]
- Xiao, Z.; Rasmann, S.; Yue, L.; Lian, F.; Zou, H.; Wang, Z. The effect of biochar amendment on N-cycling genes in soils: A meta-analysis. Sci. Total Environ. 2019, 696, 133984. [Google Scholar] [CrossRef]
- Yang, O.; Evans, S.E.; Friesen, M.L.; Tiemann, L.K. Effect of nitrogen fertilization on the abundance of nitrogen cycling genes in agricultural soils: A meta-analysis of field studies. Soil Biol. Biochem. 2018, 127, 71–78. [Google Scholar] [CrossRef]
- Wang, Q.; Ma, M.; Jiang, X.; Guan, D.; Wei, D.; Zhao, B.; Chen, S.; Cao, F.; Li, L.; Yang, X. Impact of 36 years of nitrogen fertilization on microbial community composition and soil carbon cycling-related enzyme activities in rhizospheres and bulk soils in northeast China. Appl. Soil Ecol. 2019, 136, 148–157. [Google Scholar] [CrossRef]
- Ai, C.; Liang, G.; Sun, J.; Wang, X.; He, P.; Zhou, W. Different roles of rhizosphere effect and long-term fertilization in the activity and community structure of ammonia oxidizers in a calcareous fluvo-aquic soil. Soil Biol. Biochem. 2013, 57, 30–42. [Google Scholar] [CrossRef]
- Orr, C.H.; Leifert, C.; Cummings, S.P.; Cooper, J.M. Impacts of organic and conventional crop management on diversity and activity of free-living nitrogen fixing bacteria and total bacteria are subsidiary to temporal effects. PLoS ONE 2012, 7, e52891. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jung, J.; Yeom, J.; Kim, J.; Han, J.; Lim, H.S.; Park, H. Change in gene abundance in the nitrogen biogeochemical cycle with temperature and nitrogen addition in antarctic soils. Res. Microbiol. 2011, 162, 1018–1026. [Google Scholar] [CrossRef]
- Marcia, R.R.C.; Ivanildo, E.M.; Sasha, N.J.; Clare, V.L.; Lucy, S.; Anthony, G.O. Molecular detection and quantification of nifH gene sequences in the rhizosphere of sorghum (sorghum bicolor) sown with two levels of nitrogen fertilizer. Appl. Soil Ecol. 2009, 42, 48–53. [Google Scholar]
- Juraeva, D.; George, E.; Davranov, K.; Ruppel, S. Detection and quantification of the nifH gene in shoot and root of cucumber plants. Can. J. Microbiol. 2006, 52, 731–739. [Google Scholar] [CrossRef]
- Coelho, M.R.R.; de Vos, M.; Carneiro, N.P.; Marriel, I.E.; Paiva, E.; Seldin, L. Diversity of nifH gene pools in the rhizosphere of two cultivars of sorghum (Sorghum bicolor) treated with contrasting levels of nitrogen fertilizer. FEMS Microbiol. Lett. 2008, 279, 15–22. [Google Scholar] [CrossRef] [Green Version]
- Gao, J.L.; Song, Y.Y.; Song, C.C.; Zhang, H.; Tan, W.W.; Du, Y. Effects of different concentrations of nitrogen additions on fine root morphological characteristics in permafrost peatland in the Daxing’an Mountains. Acta Ecol. Sinica 2021, 41, 8171–8177. [Google Scholar]
- Tian, J.; Dungait, J.A.J.; Lu, X.K.; Yang, Y.F.; Hartley, I.P.; Zhang, W.; Mo, J.M.; Yu, G.R.; Zhou, J.Z.; Kuzyakov, Y. Long-term nitrogen addition modifies microbial composition and functions for slow carbon cycling and increased sequestration in tropical forest soil. Glob. Change Biol. 2019, 25, 3267–3281. [Google Scholar] [CrossRef] [Green Version]
- Zhang, W.; Zhu, X.; Liu, L.; Fu, S.; Chen, H. Large difference of inhibitive effect of nitrogen deposition on soil methane oxidation between plantations with N-fixing tree species and non-N-fixing tree species. J. Geophys Res.-Biogeo. 2012, 117, G00N16. [Google Scholar] [CrossRef]
- Drabek, O.; Boruvka, L.; Mladkova, L.; Kocarek, M. Possible method of aluminium speciation in forest soils. J. Inorg. Biochem. 2003, 97, 8–15. [Google Scholar] [CrossRef]
- Zhang, Y.; Loreau, M.; He, N.; Zhang, G.; Han, X. Mowing exacerbates the loss of ecosystem stability under nitrogen enrichment in a temperate grassland. Funct. Ecol. 2017, 31, 1637–1646. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Freedman, Z.B.; Romanowicz, K.J.; Upchurch, R.A.; Zak, D.R. Differential responses of total and active soil microbial communities to long-term experimental N deposition. Soil Biol. Biochem. 2015, 90, 275–282. [Google Scholar] [CrossRef] [Green Version]
- Turlapati, S.A.; Minocha, R.; Bhiravarasa, P.S.; Tisa, L.S.; Thomas, W.K.; Minocha, S.C. Chronic N-amended soils exhibit an altered bacterial community structure in Harvard Forest, MA, USA. FEMS Microbiol. Ecol. 2012, 83, 478–493. [Google Scholar] [CrossRef] [Green Version]
- Cui, J.Y.; Yuan, X.C.; Zhang, Q.F.; Zhou, J.C.; Lin, K.M.; Xu, J.G.; Zeng, Y.Z.; Wu, Y.; Cheng, L.; Zeng, Q.X.; et al. Nutrient availability is a dominant predictor of soil bacterial and fungal community composition after nitrogen addition in subtropical acidic forests. PLoS ONE 2021, 16, e0246263. [Google Scholar] [CrossRef]
- Craine, J.M.; Morrow, C.; Fierer, N. Microbial nitrogen limitation increases decomposition. Ecology 2007, 88, 2105–2113. [Google Scholar] [CrossRef]
- Zhu, X.C.; Sun, L.Y.; Song, F.B.; Liu, S.Q.; Liu, F.L.; Li, X.N. Soil microbial community and activity are affected by integrated agricultural practices in China. Eur. J. Soil Sci. 2018, 69, 924–935. [Google Scholar] [CrossRef]
- Lu, G.R.; Xie, B.H.; Cagle, G.A.; Wang, X.H.; Han, G.X.; Wang, X.J.; Hou, A.X.; Guan, B. Effects of simulated nitrogen deposition on soil microbial community diversity in coastal wetland of the Yellow River Delta. Sci. Total Environ. 2021, 757, 143825. [Google Scholar] [CrossRef]
- Li, Y.B.; Bezemer, T.M.; Yang, J.J.; Lü, X.T.; Li, X.Y.; Liang, W.J.; Han, X.G.; Li, Q. Changes in litter quality induced by N deposition alter soil microbial communities. Soil Biol. Biochem. 2019, 130, 33–42. [Google Scholar] [CrossRef]
- Freitag, T.E.; Chang, L.; Clegg, C.D.; Prosser, J.I. Influence of inorganic nitrogen management regime on the diversity of nitrite-oxidizing bacteria in agricultural grassland soils. Appl. Environ. Microbiol. 2005, 71, 8323–8334. [Google Scholar] [CrossRef] [Green Version]
- Frey, S.D.; Knorr, M.; Parrent, J.L.; Simpson, R.T. Chronic nitrogen enrichment affects the structure and function of the soil microbial community in temperate hardwood and pine forests. For. Ecol. Manag. 2004, 196, 159–171. [Google Scholar] [CrossRef]
- Shi, X.Z.; Hu, H.W.; Wang, J.; He, J.Z.; Zheng, C.; Wan, X.; Huang, Z. Niche separation of comammox Nitrospira and canonical ammonia oxidizers in an acidic subtropical forest soil under long-term nitrogen deposition. Soil Biol. Biochem. 2018, 126, 114–122. [Google Scholar] [CrossRef]
- Kirchmann, H.; Schön, M.; Börjesson, G.; Hamnér, K.; Kätterer, T. Properties of soils in the Swedish long-term fertility experiments: VII. Changes in topsoil and upper subsoil at Örja and Fors after 50 years of nitrogen fertilization and manure application. Acta Agric. Scand. Sect. B–Soil Plant Sci. 2013, 63, 25–36. [Google Scholar] [CrossRef]
- Geisseler, D.; Scow, K.M. Long-term effects of mineral fertilizers on soil microorganisms–A review. Soil Biol. Biochem. 2014, 75, 54–63. [Google Scholar] [CrossRef]
- Yang, Y.; Cheng, H.; Gao, H.; An, S.S. Response and driving factors of soil microbial diversity related to global nitrogen addition. Land Degrad. Dev. 2020, 31, 190–204. [Google Scholar] [CrossRef]
- Zeglin, L.H.; Stursova, M.; Sinsabaugh, R.L.; Collins, S.L. Microbial responses to nitrogen addition in three contrasting grassland ecosystems. Oecologia 2007, 154, 349–359. [Google Scholar] [CrossRef]
- Song, Y.; Song, C.; Meng, H.; Swarzenski, C.M.; Wang, X.; Tan, W. Nitrogen additions affect litter quality and soil biochemical properties in a peatland of northeast china. Ecol. Eng. 2017, 100, 175–185. [Google Scholar] [CrossRef]
- Fang, H.J.; Cheng, S.L.; Yu, G.R.; Xu, M.J.; Wang, Y.S.; Li, L.S.; Dang, X.S.; Wang, L.; Li, Y.N. Experimental nitrogen deposition alters the quantity and quality of soil dissolved organic carbon in an alpine meadow on the Qinghai-Tibetan Plateau. Appl. Soil Ecol. 2014, 81, 1–11. [Google Scholar] [CrossRef]
- Deng, L.; Huang, C.; Kim, D.; Shangguan, Z.; Wang, K.; Song, X.; Peng, C. Soil GHG fluxes are altered by N deposition: New data indicate lower N stimulation of the N2O flux and greater stimulation of the calculated C pools. Glob. Chang. Biol. 2020, 26, 2613–2629. [Google Scholar] [CrossRef] [Green Version]
- Guan, B.; Xie, B.H.; Yang, S.S.; Hou, A.X.; Chen, M.; Han, G.X. Effects of five years’ nitrogen deposition on soil properties and plant growth in a salinized reed wetland of the Yellow River Delta. Ecol. Eng. 2019, 136, 160–166. [Google Scholar] [CrossRef]
- Song, C.; Liu, D.; Yang, G.; Song, Y.; Mao, R. Effect of nitrogen addition on decomposition of Calamagrostis angustifolia litters from freshwater marshes of northeast China. Ecol. Eng. 2011, 37, 1578–1582. [Google Scholar] [CrossRef]
- Jiang, C.M.; Yu, G.R.; Fang, H.J.; Cao, G.M.; Li, Y.N. Short-term effect of increasing nitrogen deposition on CO2, CH4 and N2O fluxes in an alpine meadow on the Qinghai-Tibetan Plateau, China. Atmos. Environ. 2010, 44, 2920–2926. [Google Scholar] [CrossRef]
- Zhou, J.; Guan, D.; Zhou, B.; Zhao, B.; Ma, M.; Qin, J.; Jiang, X.; Chen, S.; Cao, F.; Shen, D.; et al. Influence of 34-years of fertilization on bacterial communities in an intensively cultivated black soil in northeast China. Soil Biol. Biochem. 2015, 90, 42–51. [Google Scholar] [CrossRef]
- Wang, Q.; Jiang, X.; Guan, D.; Wei, D.; Zhao, B.; Ma, M.; Chen, S.; Li, L.; Cao, F.; Li, J. Long-term fertilization changes bacterial diversity and bacterial communities in the maize rhizosphere of Chinese Mollisols. Appl. Soil Ecol. 2018, 125, 88–96. [Google Scholar] [CrossRef]


| TC (mg·g−1) | TN (mg·g−1) | TP (mg·g−1) | AN (mg·kg−1) | NN (mg·kg−1) | DOC (mg·kg−1) | pH | |
|---|---|---|---|---|---|---|---|
| CK | 403.23 ± 12.77a | 14.07 ± 1.08c | 1.98 ± 0.01a | 39.94 ± 9.48c | 45.75 ± 17.22ab | 421.53 ± 91.65ab | 5.73 ± 0.02a |
| N1 | 406.53 ± 2.53a | 18.28 ± 0.62b | 1.79 ± 0.29ab | 123.41 ± 1.13a | 55.06 ± 0.82a | 566.47 ± 63.89a | 5.76 ± 0.02a |
| N2 | 392.89 ± 10.55a | 24.70 ± 1.09a | 1.70 ± 0.12ab | 46.56 ± 5.76c | 53.82 ± 2.44ab | 331.00 ± 15.51b | 5.75 ± 0.20a |
| N3 | 388.57 ± 9.92a | 16.90 ± 1.22b | 1.41 ± 0.20b | 95.81 ± 10.57b | 34.01 ± 0.19b | 351.33 ± 87.57b | 5.83 ± 0.06a |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, X.; Song, Y.; Song, C.; Wang, X.; Wang, N.; Gao, S.; Cheng, X.; Liu, Z.; Gao, J.; Du, Y. Effect of Nitrogen Addition on Soil Microbial Functional Gene Abundance and Community Diversity in Permafrost Peatland. Microorganisms 2021, 9, 2498. https://doi.org/10.3390/microorganisms9122498
Ma X, Song Y, Song C, Wang X, Wang N, Gao S, Cheng X, Liu Z, Gao J, Du Y. Effect of Nitrogen Addition on Soil Microbial Functional Gene Abundance and Community Diversity in Permafrost Peatland. Microorganisms. 2021; 9(12):2498. https://doi.org/10.3390/microorganisms9122498
Chicago/Turabian StyleMa, Xiuyan, Yanyu Song, Changchun Song, Xianwei Wang, Nannan Wang, Siqi Gao, Xiaofeng Cheng, Zhendi Liu, Jinli Gao, and Yu Du. 2021. "Effect of Nitrogen Addition on Soil Microbial Functional Gene Abundance and Community Diversity in Permafrost Peatland" Microorganisms 9, no. 12: 2498. https://doi.org/10.3390/microorganisms9122498





