Responses of Soil Microbial Traits to Ground Cover in Citrus Orchards in Central China
Abstract
:1. Introduction
2. Materials and Methods
2.1. Field Experiments
2.2. Sampling and Analysis
2.3. Statistical Analysis
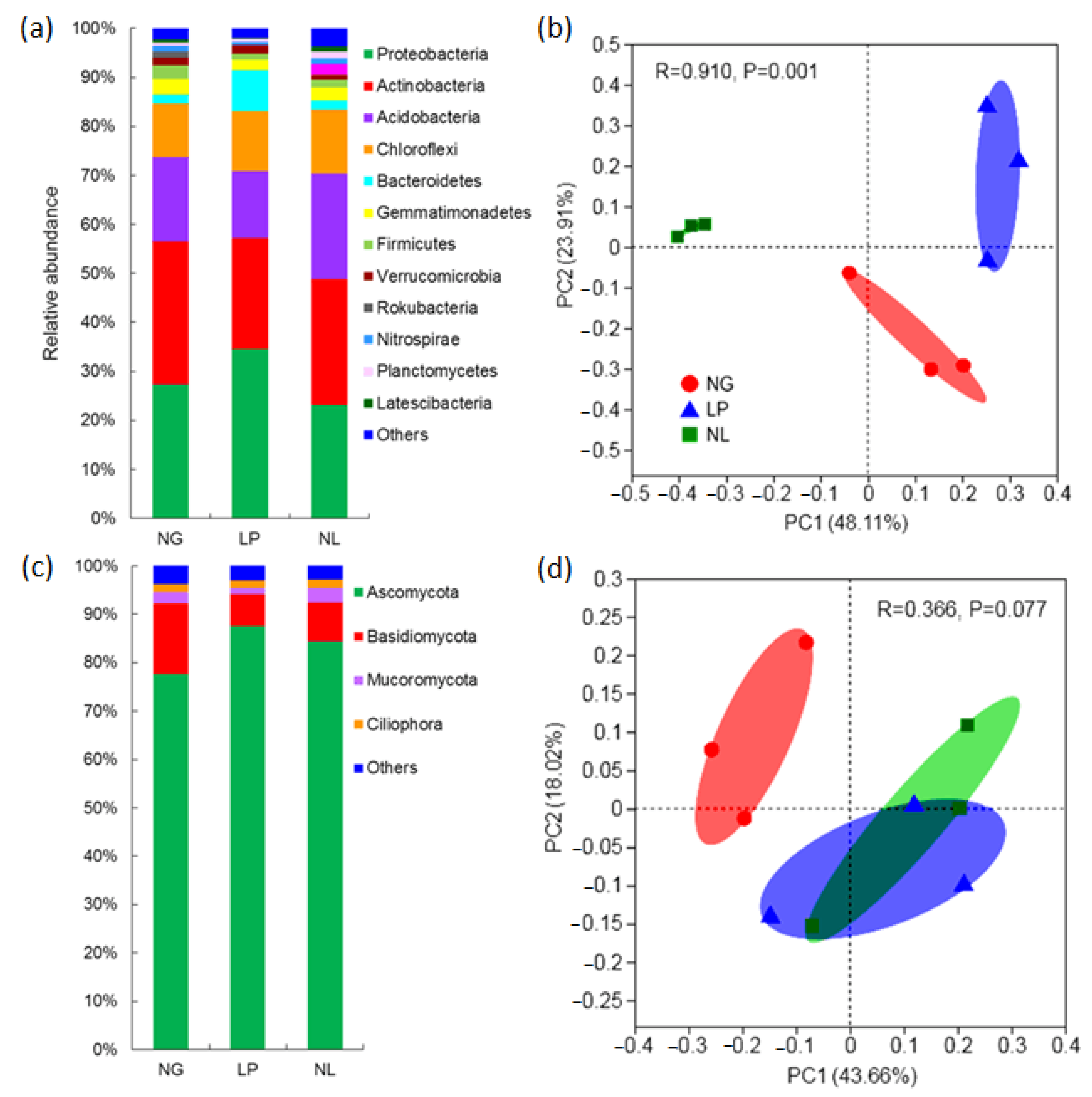
3. Results
3.1. Grasses and Soil Properties
3.2. Soil Microbial Community Composition and Diversity
3.3. Responses of Soil Microbial Community and Biomass to Grass and Soil Properties
4. Discussion
4.1. Grass Biomass Is a Strong Driver of Soil Chemical Properties
4.2. Ground Cover and Soil Properties Both Significantly Impact the Soil Microbial Community
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sanchez, E.E.; Giayetto, A.; Cichon, L.; Fernandez, D.; Aruani, M.C.; Curetti, M. Cover crops influence soil properties and tree performance in an organic apple (Malus domestica Borkh) orchard in northern Patagonia. Plant Soil 2007, 292, 193–203. [Google Scholar] [CrossRef]
- Wei, H.; Xiang, Y.Z.; Liu, Y.; Zhang, J.E. Effects of sod cultivation on soil nutrients in orchards across China: A meta-analysis. Soil Tillage Res. 2017, 169, 16–24. [Google Scholar] [CrossRef]
- Zheng, W.; Zhao, Z.Y.; Gong, Q.L.; Zhai, B.N.A.; Li, Z.Y. Effects of cover crop in an apple orchard on microbial community composition, networks, and potential genes involved with degradation of crop residues in soil. Biol. Fertil. Soils 2018, 54, 743–759. [Google Scholar] [CrossRef]
- Zheng, W.; Gong, Q.L.; Zhao, Z.Y.; Liu, J.; Zhai, B.N.; Wang, Z.H.; Li, Z.Y. Changes in the soil bacterial community structure and enzyme activities after intercrop mulch with cover crop for eight years in an orchard. Eur. J. Soil Biol. 2018, 86, 34–41. [Google Scholar] [CrossRef]
- Wu, B.; Wang, P.; Devlin, A.T.; Xiao, S.; Shu, W.; Zhang, H.; Ding, M. Influence of soil and water conservation measures on soil microbial communities in a citrus orchard of southeast China. Microorganisms 2021, 9, 319. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Z.; Huang, X.; Feng, D.; Xing, S.; Weng, B. Long-term effects of legume mulching on soil chemical properties and bacterial community composition and structure. Agric. Ecosyst. Environ. 2018, 268, 24–33. [Google Scholar] [CrossRef]
- Qian, X.; Gu, J.; Pan, H.J.; Zhang, K.Y.; Sun, W.; Wang, X.J.; Gao, H. Effects of living mulches on the soil nutrient contents, enzyme activities, and bacterial community diversities of apple orchard soils. Eur. J. Soil Biol. 2015, 70, 23–30. [Google Scholar] [CrossRef]
- Wan, X.; Huang, Z.; He, Z.; Yu, Z.; Wang, M.; Davis, M.R.; Yang, Y. Soil C:N ratio is the major determinant of soil microbial community structure in subtropical coniferous and broadleaf forest plantations. Plant Soil 2015, 387, 103–116. [Google Scholar] [CrossRef]
- Masayuki, U.; Rota, W.; Teri, C.B.; Kanehiro, K. Variations in the soil microbial community composition of a tropical montane forest ecosystem: Does tree species matter? Soil Biol. Biochem. 2008, 40, 2699–2702. [Google Scholar]
- Waldrop, M.P.; Firestone, M.K. Response of Microbial Community Composition and Function to Soil Climate Change. Microb. Ecol. 2006, 52, 716–724. [Google Scholar] [CrossRef] [PubMed]
- Wei, H.; Zhang, K.; Zhang, J.; Li, D.F.; Zhang, Y.; Xiang, H.M. Grass cultivation alters soil organic carbon fractions in a subtropical orchard of southern China. Soil Tillage Res. 2018, 181, 110–116. [Google Scholar] [CrossRef]
- Duan, J.; Liu, Y.J.; Yang, J.; Tang, C.J.; Shi, Z.H. Role of groundcover management in controlling soil erosion under extreme rainfall in citrus orchards of southern China. J. Hydrol. 2020, 582, 124290. [Google Scholar] [CrossRef]
- Cruz, A.F.; Pires, M.D.; Nascimento, L.K.B.D.; Lucrecia, M.; Andreote, F.D. Cover cropping system and mulching can shape soil microbial status in fruit orchards. Sci. Agric. 2020, 77, e20180316. [Google Scholar] [CrossRef] [Green Version]
- Zhao, J. Legume-soil interactions: Legume addition enhances the complexity of the soil food web. Plant Soil 2014, 385, 273–286. [Google Scholar] [CrossRef]
- Gastine, A.; Scherer-Lorenzen, M.; Leadley, P.W. No consistent effects of plant diversity on root biomass, soil biota and soil abiotic conditions in temperate grassland communities. Appl. Soil Ecol. 2003, 24, 101–111. [Google Scholar] [CrossRef]
- Wardle, D.A.; Yeates, G.W.; Williamson, W.; Bonner, K.I. The response of a three trophic level soil food web to the identity and diversity of plant species and functional groups. Oikos 2003, 102, 45–56. [Google Scholar] [CrossRef]
- Deyn, G.B.D.; Raaijmakers, C.E.; Ruijven, J.V.; Berendse, F.; Putten, W.H. Plant species identity and diversity effects on different trophic levels of nematodes in the soil food web. Oikos 2004, 106, 576–586. [Google Scholar] [CrossRef]
- Spehn, E.M.; Joshi, J.; Schmid, B.; Alphei, J.; Körner, C. Plant diversity effects on soil heterotrophic activity in experimental grassland ecosystems. Plant Soil 2000, 224, 217–230. [Google Scholar] [CrossRef]
- Boddey, R.M.; Peoples, M.B.; Palmer, B.; Dart, P.J. Use of the 15N natural abundance technique to quantify biological nitrogen fixation by woody perennials. Nutr. Cycl. Agroecosyst. 2000, 57, 235–270. [Google Scholar] [CrossRef]
- Vance, E.D.; Brookes, P.C.; Jenkinson, D.S. Microbial biomass measurements in forest soils: The use of the chloroform fumigation-incubation method in strongly acid soils. Soil Biol. Biochem. 1987, 19, 697–702. [Google Scholar] [CrossRef]
- Xu, N.; Tan, G.C.; Wang, H.Y.; Gai, X.P. Effect of biochar additions to soil on nitrogen leaching, microbial biomass and bacterial community structure. Eur. J. Soil Biol. 2016, 74, 1–8. [Google Scholar] [CrossRef]
- Rousk, J.; Baath, E.; Brookes, P.C.; Lauber, C.L.; Lozupone, C.; Caporaso, J.G.; Knight, R.; Fierer, N. Soil bacterial and fungal communities across a pH gradient in an arable soil. ISME J. 2010, 4, 1340–1351. [Google Scholar] [CrossRef] [PubMed]
- MacNally, R. Hierarchical partitioning as an interpretative tool in multivariate inference. Aust. J. Ecol. 1996, 21, 224–228. [Google Scholar]
- Wang, P.; Wang, Y.; Wu, Q.S. Effects of soil tillage and planting grass on arbuscular mycorrhizal fungal propagules and soil properties in citrus orchards in southeast China. Soil Tillage Res. 2016, 155, 54–61. [Google Scholar] [CrossRef]
- Sarto, M.V.M.; Borges, W.L.B.; Sarto, J.R.W.; Pires, C.A.B.; Rice, C.W.; Rosolem, C.A. Soil microbial community and activity in a tropical integrated crop-livestock system. Appl. Soil Ecol. 2020, 145, 103350. [Google Scholar] [CrossRef]
- Li, Q.; Song, Y.; Li, G.; Yu, P.; Wang, P.; Zhou, D. Grass-legume mixtures impact soil N, species recruitment, and productivity in temperate steppe grassland. Plant Soil 2015, 394, 271–285. [Google Scholar] [CrossRef]
- Paynel, F.; Lesuffleur, F.; Bigot, J.; Diquélou, S.; Cliquet, J.B. A study of 15N transfer between legumes and grasses. Agron. Sustain. Dev. 2008, 28, 281–290. [Google Scholar] [CrossRef]
- Trannin, W.S.; Urquiaga, S.; Guerra, G.; Ibijbijen, J.; Cadisch, G. Interspecies competition and N transfer in a tropical grass-legume mixture. Biol. Fertil. Soils 2000, 32, 441–448. [Google Scholar] [CrossRef]
- Schipanski, M.E.; Drinkwater, L.E. Nitrogen fixation in annual and perennial legume-grass mixtures across a fertility gradient. Plant Soil 2012, 357, 147–159. [Google Scholar] [CrossRef]
- Jensen, E.S. Grain yield, symbiotic N2 fixation and interspecific competition for inorganic N in pea-barley intercrops. Plant Soil 1996, 182, 25–38. [Google Scholar] [CrossRef]
- Fu, Y.; Lu, Y.; Heitman, J.; Ren, T. Root influences on soil bulk density measurements with thermo-time domain reflectometry. Geoderma 2021, 403, 115195. [Google Scholar] [CrossRef]
- Breulmann, M.; Schulz, E.; Weisshuhn, K.; Buscot, F. Impact of the plant community composition on labile soil organic carbon, soil microbial activity and community structure in semi-natural grassland ecosystems of different productivity. Plant Soil 2012, 352, 253–265. [Google Scholar] [CrossRef]
- Stephan, A. Meyer, A.H. Schmid, B. Plant diversity affects culturable soil bacteria in experimental grassland communities. J. Ecol. 2000, 88, 988–998. [Google Scholar] [CrossRef]
- Moore, J.M.; Klose, S.; Tabatabai, M.A. Soil microbial biomass carbon and nitrogen as affected by cropping systems. Biol. Fertil. Soils 2000, 31, 200–210. [Google Scholar] [CrossRef]
- Fox, A.; Lüscher, A.; Widmer, F. Plant species identity drives soil microbial community structures that persist under a following crop. Ecol. Evol. 2020, 10, 8652–8668. [Google Scholar] [CrossRef]
- Hammelehle, A.; Oberson, A.; Lüscher, A.; Mäder, P.; Mayer, J. Above- and belowground nitrogen distribution of a red clover-perennial ryegrass sward along a soil nutrient availability gradient established by organic and conventional cropping systems. Plant Soil 2018, 425, 507–525. [Google Scholar] [CrossRef]
- Li, C.; Yan, K.; Tang, L.; Jia, Z.; Li, Y. Change in deep soil microbial communities due to long-term fertilization. Soil Biol. Biochem. 2014, 75, 264–272. [Google Scholar] [CrossRef]
- Fierer, N.; Bradford, M.A.; Jackson, R.B. Toward an ecological classification of soil bacteria. Ecology 2007, 88, 1354–1364. [Google Scholar] [CrossRef] [PubMed]
- Toda, M.; Uchida, Y. Long-term use of green manure legume and chemical fertiliser affect soil bacterial community structures but not the rate of soil nitrate decrease when excess carbon and nitrogen are applied. Soil Res. 2017, 55, 524–533. [Google Scholar] [CrossRef]
- Weber, C.F.; Vilgalys, R.; Kuske, C.R. Changes in Fungal Community Composition in Response to Elevated Atmospheric CO2 and Nitrogen Fertilization Varies with Soil Horizon. Front. Microbiol. 2013, 4, 78. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xue, P.P.; Yolima, C.; Vanessa, P.; Budiman, M.; Mcbratney, A.B. Soil properties drive microbial community structure in a large scale transect in south eastern Australia. Sentific Rep. 2018, 8, 11725. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marschner, P.; Yang, C.H.; Lieberei, R.; Crowley, D.E. Soil and plant specific effects on bacterial community composition in the rhizosphere. Soil Biol. Biochem. 2001, 33, 1437–1445. [Google Scholar] [CrossRef]
- Potthoff, M.; Steenwerth, K.L.; Jackson, L.E.; Drenovsky, R.E.; Scow, K.M.; Joergensen, R.G. Soil microbial community composition as affected by restoration practices in California grassland. Soil Biol. Biochem. 2006, 38, 1851–1860. [Google Scholar] [CrossRef] [Green Version]



| Properties | NG | LP | NL |
|---|---|---|---|
| Dominant species (coverage) | Oxalis corniculata (25%) Duchesnea indica (25%) Veronica didyma (11%) Echinochloa crusgalli (6%) | Vicia villosa var. glabrescens (80%) Duchesnea indica (12%) Galium aparine (8%) | Vicia villosa var. glabrescens (30%) Echinochloa crusgalli (8%) Duchesnea indica (7%) Galium aparine (7%) |
| Grass coverage (%) | 71 b | 94 a | 89 a |
| SR | 13 a | 6 b | 6 b |
| SB (kg m−2) | 0.24 b | 0.32 a | 0.36 a |
| RB (kg m−2) | 0.17 c | 0.24 b | 0.33 a |
| BC (g m−2) | 170.08 c | 230.71 b | 278.19 a |
| BN (g m−2) | 8.72 b | 15.10 a | 16.42 a |
| FixN (g m−2) | 0 c | 5.71 a | 5.01 a |
| Soil Property | NG | LP | NL |
|---|---|---|---|
| BD (g cm−3) | 1.44 a | 1.42 a | 1.42 a |
| pH | 6.03 a | 6.08 a | 6.13 a |
| TC (g kg−1) | 5.09 c | 9.84 b | 13.33 a |
| TN (g kg−1) | 0.83 b | 1.26 a | 1.22 a |
| AN (mg kg−1) | 1.97 c | 2.91 a | 2.28 b |
| NN (mg kg−1) | 15.02 b | 19.32 a | 20.16 a |
| DOC (mg kg−1) | 13.73 b | 21.12 a | 25.63 a |
| DON (mg kg−1) | 26.09 b | 36.04 ab | 47.88 a |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, Y.; Wang, X.; Hu, R.; Zhao, J.; Jiang, Y. Responses of Soil Microbial Traits to Ground Cover in Citrus Orchards in Central China. Microorganisms 2021, 9, 2507. https://doi.org/10.3390/microorganisms9122507
Wu Y, Wang X, Hu R, Zhao J, Jiang Y. Responses of Soil Microbial Traits to Ground Cover in Citrus Orchards in Central China. Microorganisms. 2021; 9(12):2507. https://doi.org/10.3390/microorganisms9122507
Chicago/Turabian StyleWu, Yupeng, Xue Wang, Ronggui Hu, Jinsong Zhao, and Yanbin Jiang. 2021. "Responses of Soil Microbial Traits to Ground Cover in Citrus Orchards in Central China" Microorganisms 9, no. 12: 2507. https://doi.org/10.3390/microorganisms9122507
APA StyleWu, Y., Wang, X., Hu, R., Zhao, J., & Jiang, Y. (2021). Responses of Soil Microbial Traits to Ground Cover in Citrus Orchards in Central China. Microorganisms, 9(12), 2507. https://doi.org/10.3390/microorganisms9122507






