Desulfovibrio desulfuricans AY5 Isolated from a Patient with Autism Spectrum Disorder Binds Iron in Low-Soluble Greigite and Pyrite
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strain AY5 Cultivation and Physiological Tests
2.2. Strain AY5 Genome Sequencing
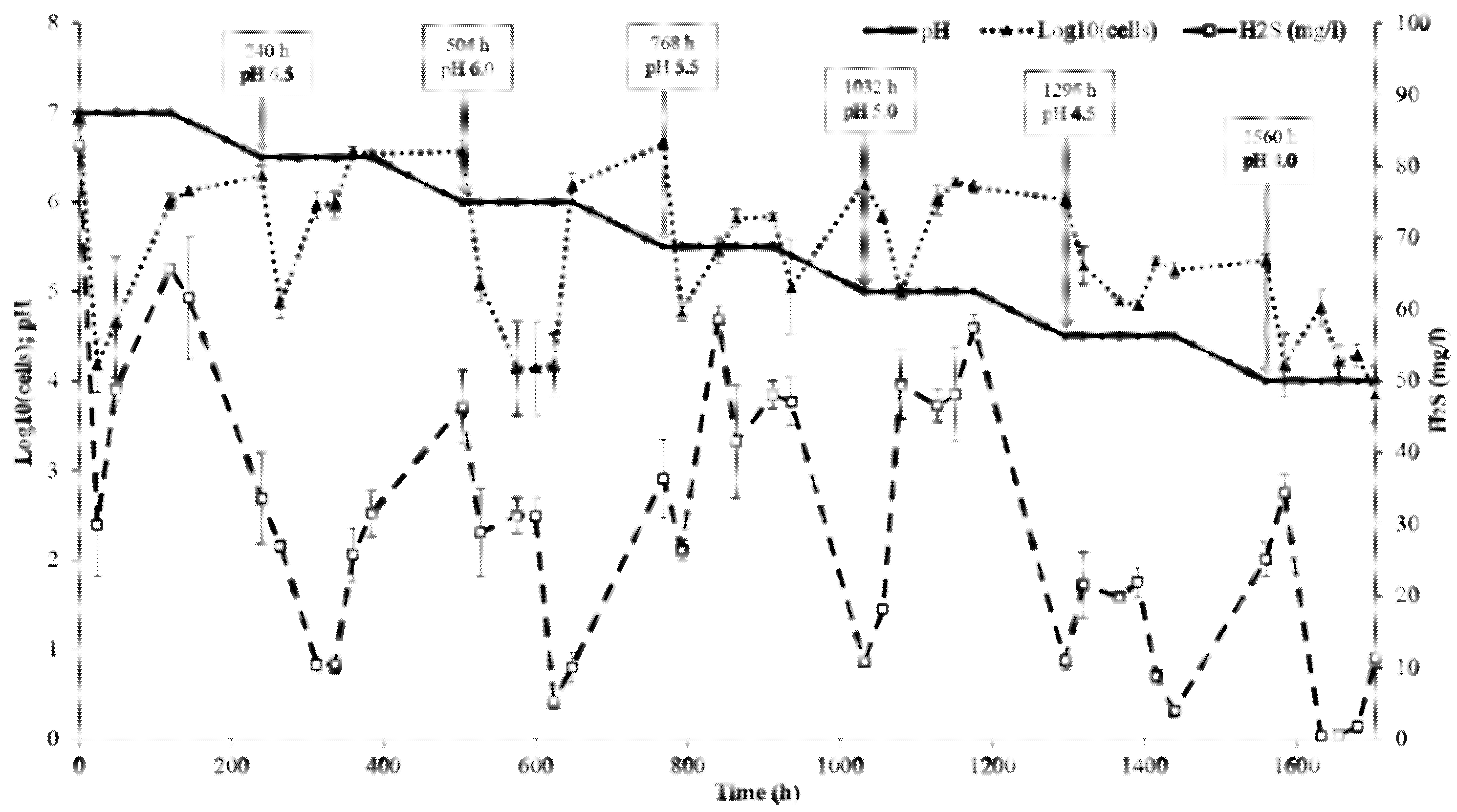
2.3. Bioreactor Culture and pH Gradient
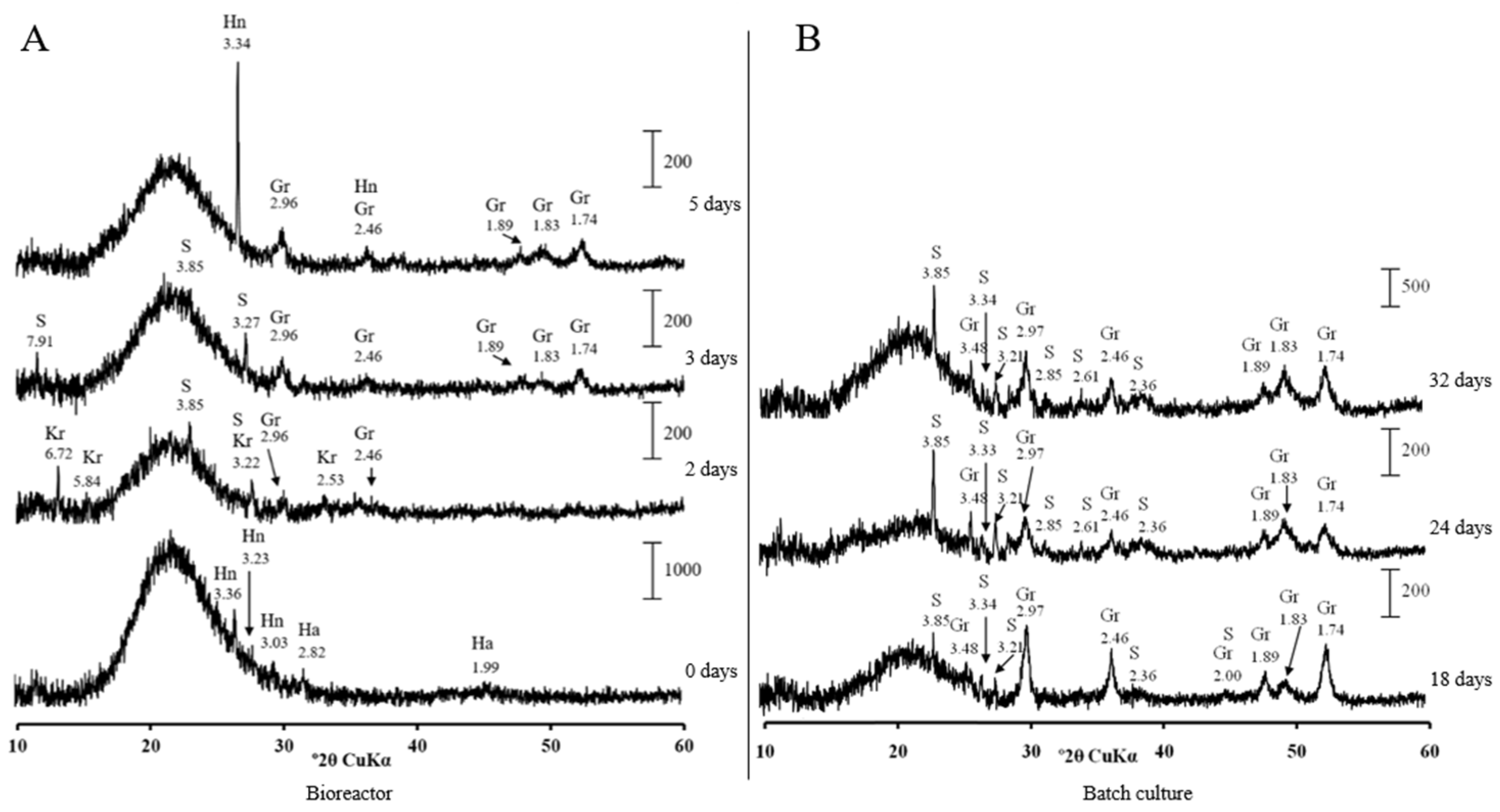
2.4. XRD
3. Results
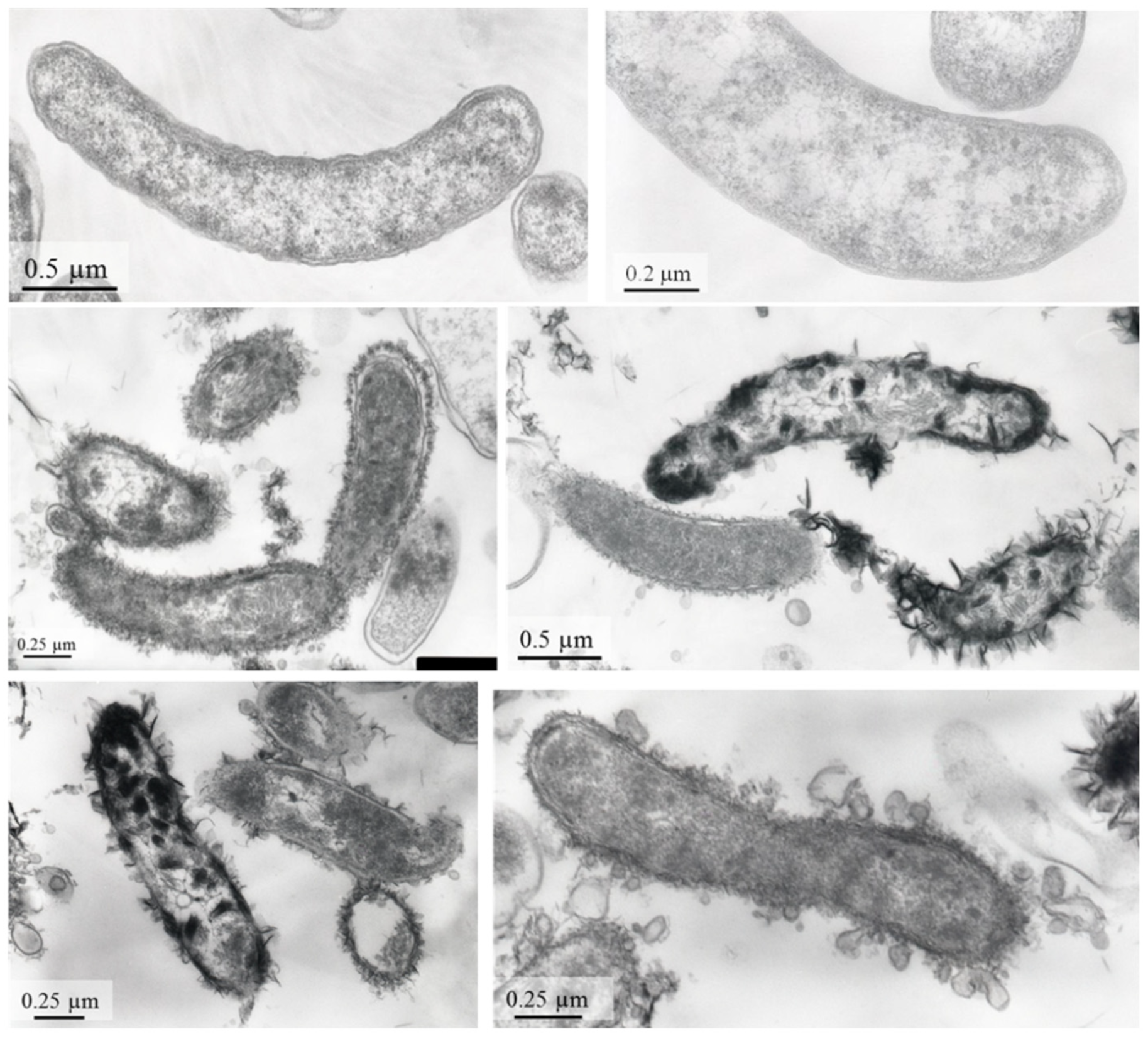
3.1. Strain AY5 Genome and Physiological Properties
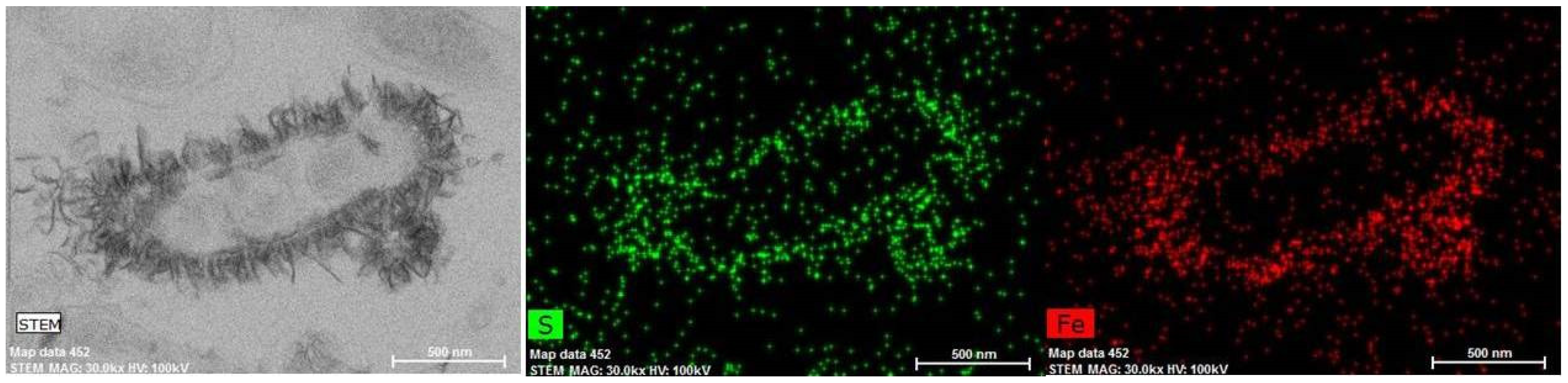
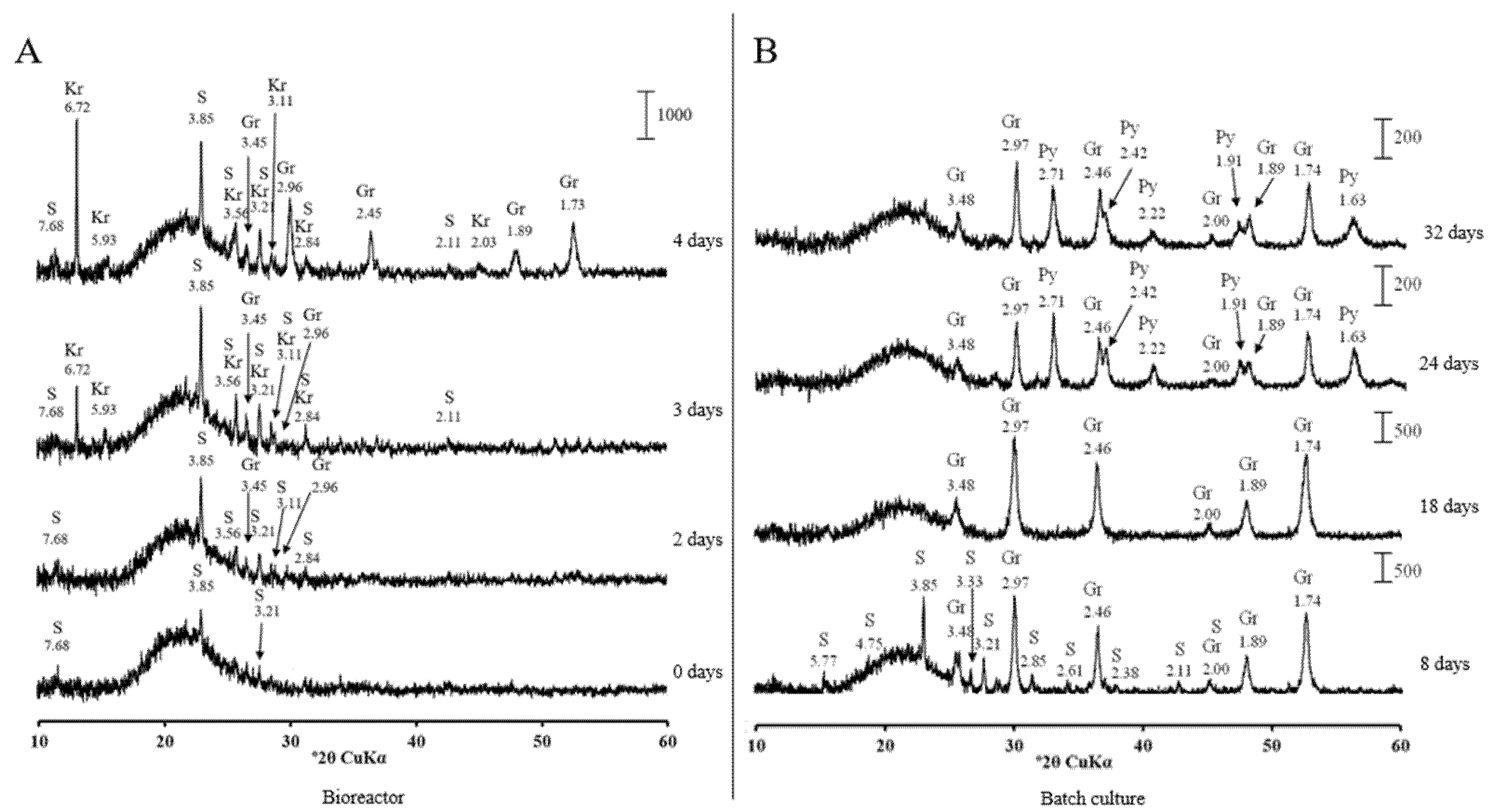
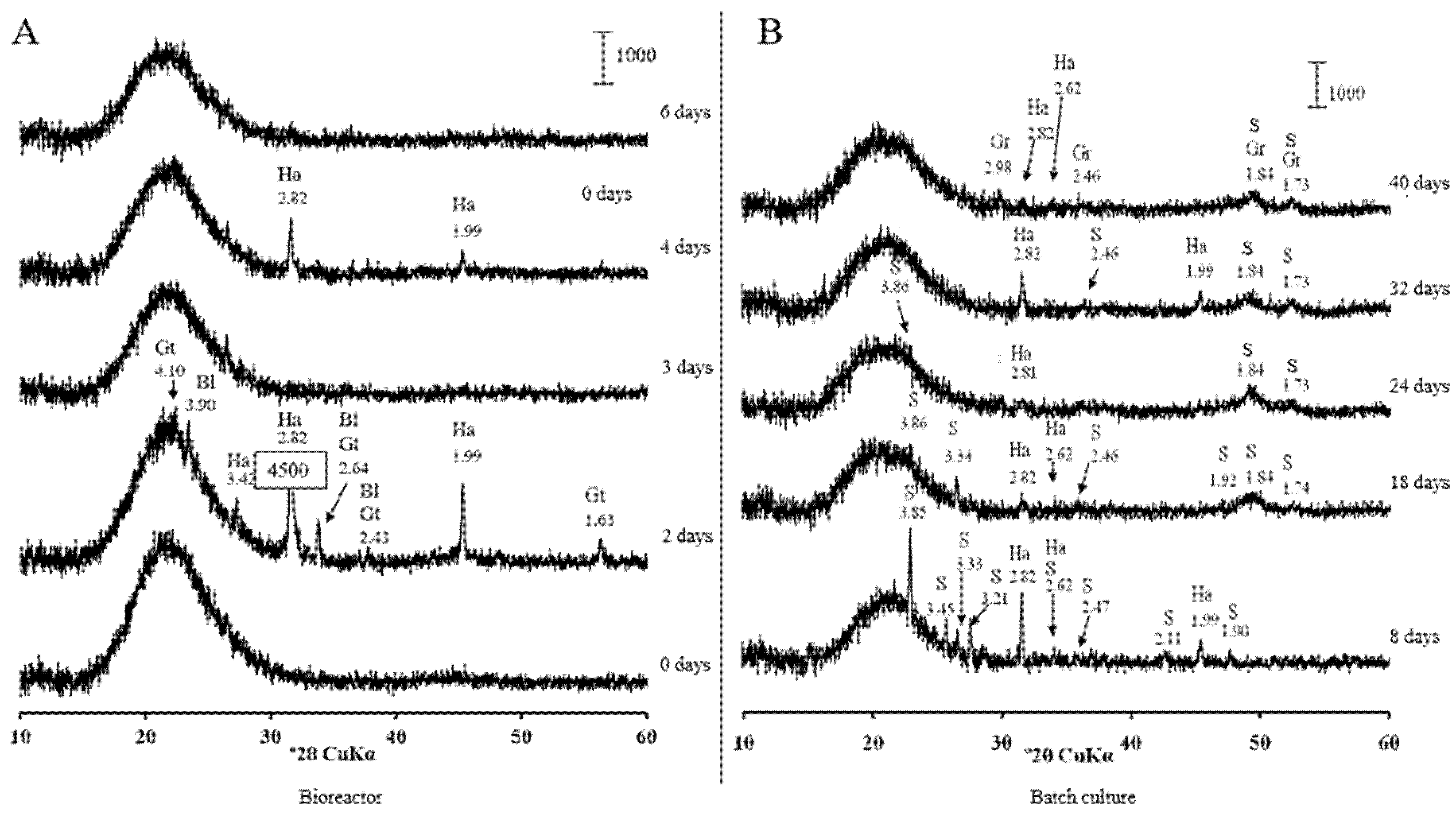
3.2. Low-Soluble Sulphide Production by Strain AY5 at Different pH
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Catalá-López, F.; Ridao, M.; Hurtado, I.; Núñez-Beltrán, A.; Gènova-Maleras, R.; Alonso-Arroyo, A.; Tobías, A.; Aleixandre-Benavent, R.; Catalá, M.A.; Tabarés-Seisdedos, R. Prevalence and Comorbidity of Autism Spectrum Disorder in Spain: Study Protocol for a Systematic Review and Meta-Analysis of Observational Studies. Syst. Rev. 2019, 8, 141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- GBD 2016 Disease and Injury Incidence and Prevalence Collaborators. Global, Regional, and National Incidence, Prevalence, and Years Lived with Disability for 328 Diseases and Injuries for 195 Countries, 1990–2016: A Systematic Analysis for the Global Burden of Disease Study 2016. Lancet 2017, 390, 1211–1259. [Google Scholar] [CrossRef] [Green Version]
- Srikantha, P.; Mohajeri, M.H. The Possible Role of the Microbiota-Gut-Brain-Axis in Autism Spectrum Disorder. Int. J. Mol. Sci. 2019, 20, 2115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bezawada, N.; Phang, T.H.; Hold, G.L.; Hansen, R. Autism Spectrum Disorder and the Gut Microbiota in Children: A Systematic Review. Ann. Nutr. Metab. 2020, 76, 16–29. [Google Scholar] [CrossRef] [PubMed]
- Johnson, D.; Letchumanan, V.; Thurairajasingam, S.; Lee, L.H. A Revolutionizing Approach to Autism Spectrum Disorder using the Microbiome. Nutrients 2020, 12, 1983. [Google Scholar] [CrossRef] [PubMed]
- Kang, D.W.; Adams, J.B.; Coleman, D.M.; Pollard, E.L.; Maldonado, J.; McDonough-Means, S.; Caporaso, J.G.; Krajmalnik-Brown, R. Long-Term Benefit of Microbiota Transfer Therapy on Autism Symptoms and Gut Microbiota. Sci. Rep. 2019, 9, 5821. [Google Scholar] [CrossRef] [PubMed]
- Finegold, S.M. Desulfovibrio Species Are Potentially Important in Regressive Autism. Med. Hypotheses 2011, 77, 270–274. [Google Scholar] [CrossRef]
- Finegold, S.M.; Downes, J.; Summanen, P.H. Microbiology of Regressive Autism. Anaerobe 2012, 18, 260–262. [Google Scholar] [CrossRef]
- De Angelis, M.; Piccolo, M.; Vannini, L.; Siragusa, S.; De Giacomo, A.; Serrazzanetti, D.I.; Cristofori, F.; Guerzoni, M.E.; Gobbetti, M.; Francavilla, R. Fecal Microbiota and Metabolome of Children with Autism and Pervasive Developmental Disorder Not Otherwise Specified. PLoS ONE 2013, 8, e76993. [Google Scholar] [CrossRef] [Green Version]
- Liu, F.; Li, J.; Wu, F.; Zheng, H.; Peng, Q.; Zhou, H. Altered Composition and Function of Intestinal Microbiota in Autism Spectrum Disorders: A Systematic Review. Transl. Psychiatry 2019, 9, 43. [Google Scholar] [CrossRef]
- Tomova, A.; Husarova, V.; Lakatosova, S.; Bakos, J.; Vlkova, B.; Babinska, K.; Ostatnikova, D. Gastrointestinal Microbiota in Children with Autism in Slovakia. Physiol. Behav. 2015, 138, 179–187. [Google Scholar] [CrossRef]
- Ding, H.T.; Taur, Y.; Walkup, J.T. Gut Microbiota and Autism: Key Concepts and Findings. J. Autism Dev. Disord. 2017, 47, 480–489. [Google Scholar] [CrossRef]
- Sidrak, S.; Yoong, T.; Woolfenden, S. Iron Deficiency in Children with Global Developmental Delay and Autism Spectrum Disorder. J. Paediatr. Child Health 2014, 50, 356–361. [Google Scholar] [CrossRef]
- Yanagimoto, Y.; Ishizaki, Y.; Kaneko, K. Iron Deficiency Anemia, Stunted Growth, and Developmental Delay Due to Avoidant/Restrictive Food Intake Disorder by Restricted Eating in Autism Spectrum Disorder. Biopsychosoc. Med. 2020, 14, 8. [Google Scholar] [CrossRef]
- Rickard, D.; Morse, J.W. Acid Volatile Sulfide (AVS). Mar. Chem. 2005, 97, 141–197. [Google Scholar] [CrossRef]
- Rickard, D. The Microbiological Formation of Iron Sulphides. Stockh. Contrib. Geology 1969, 20, 49–66. [Google Scholar]
- Neal, A.L.; Techkarnjanaruk, S.; Dohnalkova, A.; McCready, D.; Peyton, B.M.; Geesey, G.G. Iron Sulfides and Sulfur Species Produced at Hematite Surfaces in the Presence of Sulfate-Reducing Bacteria. Geochim. Cosmochim. Acta 2001, 65, 223–235. [Google Scholar] [CrossRef]
- Smith, E.A.; Macfarlane, G.T. Enumeration of Amino Acid Fermenting Bacteria in the Human Large Intestine: Effects of pH and Starch on Peptide Metabolism and Dissimilation of Amino Acids. FEMS Microbiol. Ecol. 1998, 25, 355–368. [Google Scholar] [CrossRef]
- Koziolek, M.; Grimm, M.; Becker, D.; Iordanov, V.; Zou, H.; Shimizu, J.; Wanke, C.; Garbacz, G.; Weitschies, W. Investigation of pH and Temperature Profiles in the GI Tract of Fasted Human Subjects Using the Intellicap® System. J. Pharm. Sci. 2015, 104, 2855–2863. [Google Scholar] [CrossRef]
- Wetzel, D.; McBride, S.M. The Impact of pH on Clostridioides Difficile Sporulation and Physiology. Appl. Environ. Microbiol. 2020, 86, e02706-19. [Google Scholar] [CrossRef]
- Ikkert, O.P.; Ivanov, M.V.; Ukhova, A.; Zuysman, V.S.; Glukhova, L.B.; Avakyan, M.R.; Karnachuk, O.V. Desulfovibrio Isolate from the Microbiote of Children with Autistic Spectrum Disorders Immobilizes Iron in Poorly Soluble Crystalline Sulfides. Microbiology 2021, 90, 268–271. [Google Scholar] [CrossRef]
- Widdel, F.; Bak, F. Gram Negative Mesophilic Sulfate Reducing Bacteria. In The Prokaryotes: A Handbook on the Biology of Bacteria: Ecophysiology, Isolation, Identification, Applications; Balows, A., Ed.; Springer: Berlin, Germany, 1992; pp. 3352–3378. [Google Scholar]
- Karnachuk, O.V.; Pimenov, N.V.; Iusupov, S.K.; Frank, I.A.; Puhakka, J.A.; Ivanov, M.V. Distribution, Diversity, and Activity of Sulfate-Reducing Bacteria in the Water Column in Gek-Gel Lake, Azerbaijan. Mikrobiologiia 2006, 75, 101–109. [Google Scholar] [CrossRef]
- Karnachuk, O.V.; Frank, Y.A.; Lukina, A.P.; Kadnikov, V.V.; Beletsky, A.V.; Mardanov, A.V.; Ravin, N.V. Domestication of Previously Uncultivated Candidatus DesulforudisAudaxviator from a Deep Aquifer in Siberia Sheds Light on Its Physiology and Evolution. ISME J. 2019, 13, 1947–1959. [Google Scholar] [CrossRef]
- Ikkert, O.P.; Gerasimchuk, A.L.; Bukhtiyarova, P.A.; Tuovinen, O.H.; Karnachuk, O.V. Characterization of Precipitates Formed by H2S-Producing, Cu-Resistant Firmicute Isolates of Tissierella from Human Gut and Desulfosporosinus from Mine Waste. Antonie Van Leeuwenhoek 2013, 103, 1221–1234. [Google Scholar] [CrossRef]
- Magoč, T.; Salzberg, S.L. FLASH: Fast Length Adjustment of Short Reads to Improve Genome Assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef]
- Nurk, S.; Bankevich, A.; Antipov, D.; Gurevich, A.A.; Korobeynikov, A.; Lapidus, A.; Prjibelski, A.D.; Pyshkin, A.; Sirotkin, A.; Sirotkin, Y.; et al. Assembling Single-Cell Genomes and Mini-Metagenomes from Chimeric MDA Products. J. Comput. Biol. 2013, 20, 714–737. [Google Scholar] [CrossRef] [Green Version]
- Brettin, T.; Davis, J.J.; Disz, T.; Edwards, R.A.; Gerdes, S.; Olsen, G.J.; Olson, R.; Overbeek, R.; Parrello, B.; Pusch, G.D.; et al. RASTtk: A Modular and Extensible Implementation of the RAST Algorithm for Building Custom Annotation Pipelines and Annotating Batches of Genomes. Sci. Rep. 2015, 5, 8365. [Google Scholar] [CrossRef] [Green Version]
- Parks, D.H.; Imelfort, M.; Skennerton, C.T.; Hugenholtz, P.; Tyson, G.W. CheckM: Assessing the Quality of Microbial Genomes Recovered from Isolates, Single Cells, and Metagenomes. Genome Res. 2015, 25, 1043–1055. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chaumeil, P.A.; Mussig, A.J.; Hugenholtz, P.; Parks, D.H. GTDB-Tk: A Toolkit to Classify Genomes with the Genome Taxonomy Database. Bioinformatics 2019, 36, 1925–1927. [Google Scholar] [CrossRef] [PubMed]
- Jain, C.; Rodriguez-R, L.M.; Phillippy, A.M.; Konstantinidis, K.T.; Aluru, S. High Throughput ANI Analysis of 90K Prokaryotic Genomes Reveals Clear Species Boundaries. Nat. Commun. 2018, 9, 5114. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mobley, H.L.; Island, M.D.; Hausinger, R.P. Molecular Biology of Microbial Ureases. Microbiol. Rev. 1995, 59, 451–480. [Google Scholar] [CrossRef]
- Beckers, G.; Bendt, A.K.; Krämer, R.; Burkovski, A. Molecular Identification of the Urea Uptake System and Transcriptional Analysis of Urea Transporter- and Urease-Encoding Genes in Corynebacterium Glutamicum. J. Bacteriol. 2004, 186, 7645–7652. [Google Scholar] [CrossRef] [Green Version]
- Sayavedra, L.; Li, T.; Batista, M.B.; Seah, B.K.B.; Booth, C.; Zhai, Q.; Chen, W.; Narbad, A. Desulfovibriodiazotrophicus sp. nov., a Sulfate-Reducing Bacterium from the Human Gut Capable of Nitrogen Fixation. Environ. Microbiol. 2021, 23, 3164–3181. [Google Scholar] [CrossRef]
- Righetto, R.D.; Anton, L.; Adaixo, R.; Jakob, R.P.; Zivanov, J.; Mahi, M.A.; Ringler, P.; Schwede, T.; Maier, T.; Stahlberg, H. High-resolution Cryo-EM Structure of Urease from the Pathogen Yersinia Enterocolitica. Nat. Commun. 2020, 11, 5101. [Google Scholar]
- Talarico, V.; Giancotti, L.; Mazza, G.A.; Miniero, R.; Bertini, M. Iron Deficiency Anemia in Celiac Disease. Nutrients 2021, 13, 1695. [Google Scholar] [CrossRef]
- Matamoros-Veloza, A.; Cespedes, O.; Johnson, B.R.G.; Stawski, T.M.; Terranova, U.; de Leeuw, N.H.; Benning, L.G. A Highly Reactive Precursor in the Ironsulfide System. Nat. Commun. 2018, 9, 3125. [Google Scholar] [CrossRef]
- Sicard, J.F.; Le Bihan, G.; Vogeleer, P.; Jacques, M.; Harel, J. Interactions of Intestinal Bacteria with Components of the Intestinal Mucus. Front. Cell Infect. Microbiol. 2017, 7, 387. [Google Scholar] [CrossRef]
- Croix, J.A.; Carbonero, F.; Nava, G.M.; Russell, M.; Greenberg, E.; Gaskins, H.R. On the Relationship between Sialomucin and Sulfomucin Expression and Hydrogenotrophic Microbes in the Human Colonic Mucosa. PLoS ONE 2011, 6, e24447. [Google Scholar] [CrossRef] [Green Version]
- Barton, L.L.; Ritz, N.L.; Fauque, G.D.; Lin, H.C. Sulfur Cycling and the Intestinal Microbiome. Dig. Dis. Sci. 2017, 62, 2241–2257. [Google Scholar] [CrossRef]
- Kushkevych, I.; Martínková, K.; Vítězová, M.; Rittmann, S.K.R. Intestinal Microbiota and Perspectives of the Use of Meta-Analysis for Comparison of Ulcerative Colitis Studies. J. Clin. Med. 2021, 10, 462. [Google Scholar] [CrossRef]
- Deplancke, B.; Hristova, K.R.; Oakley, H.A.; McCracken, V.J.; Aminov, R.; Mackie, R.I.; Gaskins, H.R. Molecular Ecological Analysis of the Succession and Diversity of Sulfate-Reducing Bacteria in the Mouse Gastrointestinal Tract. Appl. Environ. Microbiol. 2000, 66, 2166–2174. [Google Scholar] [CrossRef] [Green Version]
- Yap, C.X.; Henders, A.K.; Alvares, G.A.; Wood, D.L.A.; Krause, L.; Tyson, G.W.; Restuadi, R.; Wallace, L.; McLaren, T.; Hansell, N.K.; et al. Autism-Related Dietary Preferences Mediate Autism-Gut Microbiome Associations. Cell 2021, 184, 5916–5931. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Karnachuk, O.V.; Ikkert, O.P.; Avakyan, M.R.; Knyazev, Y.V.; N.Volochaev, M.; Zyusman, V.S.; Panov, V.L.; Kadnikov, V.V.; Mardanov, A.V.; Ravin, N.V. Desulfovibrio desulfuricans AY5 Isolated from a Patient with Autism Spectrum Disorder Binds Iron in Low-Soluble Greigite and Pyrite. Microorganisms 2021, 9, 2558. https://doi.org/10.3390/microorganisms9122558
Karnachuk OV, Ikkert OP, Avakyan MR, Knyazev YV, N.Volochaev M, Zyusman VS, Panov VL, Kadnikov VV, Mardanov AV, Ravin NV. Desulfovibrio desulfuricans AY5 Isolated from a Patient with Autism Spectrum Disorder Binds Iron in Low-Soluble Greigite and Pyrite. Microorganisms. 2021; 9(12):2558. https://doi.org/10.3390/microorganisms9122558
Chicago/Turabian StyleKarnachuk, Olga V., Olga P. Ikkert, Marat R. Avakyan, Yurii V. Knyazev, Mikhail N.Volochaev, Viacheslav S. Zyusman, Vasily L. Panov, Vitaly V. Kadnikov, Andrey V. Mardanov, and Nikolai V. Ravin. 2021. "Desulfovibrio desulfuricans AY5 Isolated from a Patient with Autism Spectrum Disorder Binds Iron in Low-Soluble Greigite and Pyrite" Microorganisms 9, no. 12: 2558. https://doi.org/10.3390/microorganisms9122558
APA StyleKarnachuk, O. V., Ikkert, O. P., Avakyan, M. R., Knyazev, Y. V., N.Volochaev, M., Zyusman, V. S., Panov, V. L., Kadnikov, V. V., Mardanov, A. V., & Ravin, N. V. (2021). Desulfovibrio desulfuricans AY5 Isolated from a Patient with Autism Spectrum Disorder Binds Iron in Low-Soluble Greigite and Pyrite. Microorganisms, 9(12), 2558. https://doi.org/10.3390/microorganisms9122558






