Antimicrobial Activity of Lactococcus lactis subsp. lactis Isolated from a Stranded Cuvier’s Beaked Whale (Ziphius cavirostris) against Gram-Positive and -Negative Bacteria
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection and Stranded Whale Species Identification
2.2. LAB Isolation from the Fecal Sample
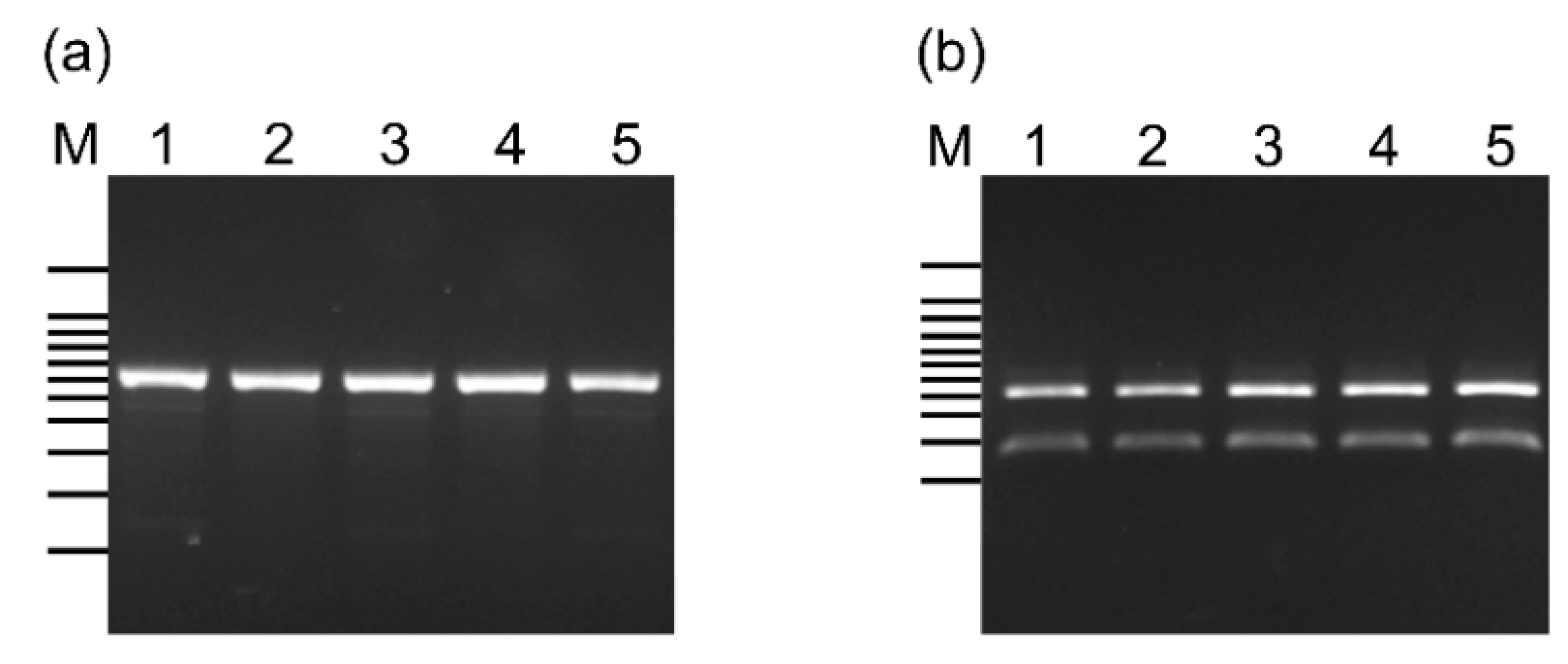
2.3. Screening for L. lactis Using Species-Specific Primers and 16S rRNA Gene Sequencing
2.4. Classification of L. lactis Subspecies Using PCR-Restriction Fragment Length Polymorphism Analysis
2.5. Random Amplified Polymorphic DNA (RAPD)-PCR Analysis
2.6. Biochemical Characterization
2.7. Antimicrobial Activity Assay
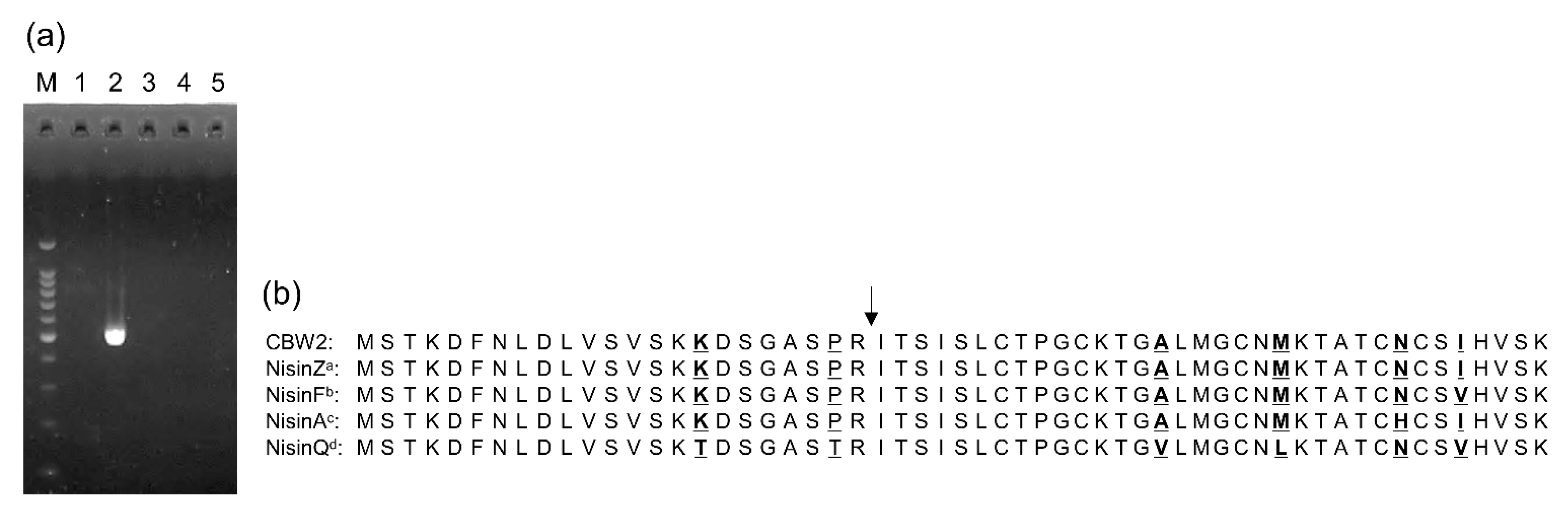
2.8. Genetical Analysis for Bacteriocin Production Potential
3. Results
3.1. Isolation of L. lactis Strains from a Stranded Cuvier’s Beaked Whale
3.2. Classification of L. lactis Subspecies in the CBW1–5 Strains
3.3. Biochemical Characterization
3.4. Antimicrobial Activity Assay
3.5. Genetic Evidence for Nisin Synthesis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Heczko, P.; Strus, M.; Kochan, P. Critical evaluation of probiotic activity. J. Physiol. Pharmacol. 2006, 57, 5–12. [Google Scholar] [PubMed]
- Guarner, F.; Khan, A.G.; Garisch, J.; Eliakim, R.; Gangl, A.; Thomson, A.; Krabshuis, J.; Lemair, T.; Kaufmann, P.; de Paula, J.A.; et al. World gastroenterology organisation global guidelines: Probiotics and prebiotics October 2011. J. Clin. Gastroenterol. 2011, 46, 468–481. [Google Scholar] [CrossRef] [PubMed]
- Fink, L.N.; Zeuthen, L.H.; Christensen, H.R.; Morandi, B.; Frøkiær, H.; Ferlazzo, G. Distinct gut-derived lactic acid bacteria elicit divergent dendritic cell-mediated NK cell responses. Int. Immunol. 2007, 19, 1319–1327. [Google Scholar] [CrossRef] [PubMed]
- Dantas, A.B.; Jesus, V.F.; Silva, R.; Almada, C.N.; Esmerino, E.A.; Cappato, L.P.; Silva, M.C.; Raices, R.S.L.; Cavalcanti, R.N.; Carvalho, C.C.; et al. Manufacture of probiotic minas frescal cheese with Lactobacillus casei Zhang. J. Dairy Sci. 2016, 99, 18–30. [Google Scholar] [CrossRef] [PubMed]
- Dal Bello, B.; Rantsiou, K.; Bellio, A.; Zeppa, G.; Ambrosoli, R.; Civera, T.; Cocolin, L. Microbial ecology of artisanal products from North West of Italy and antimicrobial activity of the autochthonous populations. LWT-Food Sci. Technol. 2010, 43, 1151–1159. [Google Scholar] [CrossRef]
- Tsuchida, S.; Kakooza, S.; Mbehang Nguema, P.P.; Wampande, E.M.; Ushida, K. Characteristics of gorilla-specific Lactobacillus isolated from captive and wild gorillas. Microorganisms 2018, 6, 86. [Google Scholar] [CrossRef] [PubMed]
- Kanamori, Y.; Hashizume, K.; Sugiyama, M.; Morotomi, M.; Yuki, N. Combination therapy with Bifidobacterium breve, Lactobacillus casei, and galactooligosaccharides dramatically improved the intestinal function in a girl with short bowel syndrome: A novel synbiotics therapy for intestinal failure. Dig. Dis. Sci. 2001, 46, 2010–2016. [Google Scholar] [CrossRef]
- Liu, Q.; Ni, X.; Wang, Q.; Peng, Z.; Niu, L.; Xie, M.; Lin, Y.; Zhou, Y.; Sun, H.; Pan, K.; et al. Investigation of lactic acid bacteria isolated from Giant Panda Feces for potential probiotics in vitro. Probiotics. Antimicrob. Proteins. 2019, 11, 85–91. [Google Scholar] [CrossRef] [PubMed]
- Klaenhammer, T.R. Genetics of bacteriocins produced by lactic acid bacteria. FEMS Microbiol. Rev. 1993, 12, 39–85. [Google Scholar] [CrossRef]
- Dobson, A.; Crispie, F.; Rea, M.C.; O’Sullivan, O.; Casey, P.G.; Lawlor, P.G.; Cotter, P.D.; Ross, P.; Gardiner, G.E.; Hill, C. Fate and efficacy of lacticin 3147-producing Lactococcus lactis in the mammalian gastrointestinal tract. FEMS Microbiol. Ecol. 2011, 76, 602–614. [Google Scholar] [CrossRef]
- Punyauppa-path, S.; Phumkhachorn, P.; Rattanachaikunsopon, P. Nisin: Production and mechanism of antimicrobial action. Int. J. Curr. Res. Rev. 2015, 7, 47. [Google Scholar]
- Schuller, F.; Benz, R.; Sahl, H.G. The peptide antibiotic subtilin acts by formation of voltage-dependent multi-state pores in bacterial and artificial membranes. Eur. J. Biochem. 1989, 182, 181–186. [Google Scholar] [CrossRef] [PubMed]
- Kuwano, K.; Tanaka, N.; Shimizu, T.; Nagatoshi, K.; Nou, S.; Sonomoto, K. Dual antibacterial mechanisms of nisin Z against Gram-positive and Gram-negative bacteria. Int. J. Antimicrob. Agents 2005, 26, 396–402. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Fang, J.; Tian, Y.; Lu, X.Y. Mechanisms of nisin resistance in Gram-positive bacteria. Ann. Microbiol. 2014, 64, 413–420. [Google Scholar] [CrossRef]
- Diaz, M.A.; Bik, E.M.; Carlin, K.P.; Venn-Watson, S.K.; Jensen, E.D.; Jones, S.E.; Gaston, E.P.; Relman, D.A.; Versalovic, J. Identification of Lactobacillus strains with probiotic features from the bottlenose dolphin (Tursiops truncatus). J. Appl. Microbiol. 2013, 115, 1037–1051. [Google Scholar] [CrossRef]
- Vela, A.I.; Fernandez, A.; los Monteros, A.E.; Goyache, J.; Herraez, P.; Tames, B.; Cruz, F.; Domínguez, L.; Fernandez-Garayzabal, J.F. Lactobacillus ceti sp. nov., isolated from beaked whales (Ziphius cavirostris). Int. J. Syst. Evol. Microbiol. 2008, 58, 891–894. [Google Scholar] [CrossRef]
- Vela, A.I.; Fernández, A.; De Quirós, Y.B.; Herráez, P.; Domínguez, L.; Fernández-Garayzábal, J.F. Weissella ceti sp. nov., isolated from beaked whales (Mesoplodon bidens). Int. J. Syst. Evol. Microbiol. 2011, 61, 2758–2762. [Google Scholar] [CrossRef]
- Evans, J.J.; Pasnik, D.J.; Klesius, P.H.; Al-Ablani, S. First report of Streptococcus agalactiae and Lactococcus garvieae from a wild bottlenose dolphin (Tursiops truncatus). J. Wildl. Dis. 2006, 42, 561–569. [Google Scholar] [CrossRef]
- Kawanishi, M.; Kojima, A.; Ishihara, K.; Esaki, H.; Kijima, M.; Takahashi, T.; Suzuki, S.; Tamura, Y. Drug resistance and pulsed-field gel electrophoresis patterns of Lactococcus garvieae isolates from cultured Seriola (yellowtail, amberjack and kingfish) in Japan. Lett. Appl. Microbiol. 2005, 40, 322–328. [Google Scholar] [CrossRef]
- Pot, B.; Devriese, L.A.; Ursi, D.; Vandamme, P.; Haesebrouck, F.; Kersters, K. Phenotypic identification and differentiation of Lactococcus strains isolated from animals. Syst. Appl. Microbiol. 1996, 19, 213–222. [Google Scholar] [CrossRef]
- Sugita, H.; Ohta, K.; Kuruma, A.; Sagesaka, T. An antibacterial effect of Lactococcus lactis isolated from the intestinal tract of the Amur catfish, Silurus asotus Linnaeus. Aquac. Res. 2007, 38, 1002–1004. [Google Scholar] [CrossRef]
- Itoi, S.; Abe, T.; Washio, S.; Ikuno, E.; Kanomata, Y.; Sugita, H. Isolation of halotolerant Lactococcus lactis subsp. lactis from intestinal tract of coastal fish. Int. J. Food Microbiol. 2008, 121, 116–121. [Google Scholar] [CrossRef] [PubMed]
- Sahnouni, F.; Boutibamaatallah, A.; Bouhadi, D.; Boutiba, Z. Characterization of bacteriocin produced by Lactococcus lactis ssp. lactis strains isolated from marine fish caught in the Algerian west coast. Türk Tarım Doğa Bilimleri Dergisi 2014, 1, 1838–1843. [Google Scholar]
- Nguyen, T.L.; Park, C.I.; Kim, D.H. Improved growth rate and disease resistance in olive flounder, Paralichthys olivaceus, by probiotic Lactococcus lactis WFLU12 isolated from wild marine fish. Aquaculture 2017, 471, 113–120. [Google Scholar] [CrossRef]
- Klijn, N.; Weerkamp, A.H.; De Vos, W.M. Genetic marking of Lactococcus lactis shows its survival in the human gastrointestinal tract. Appl. Environ. Microbiol. 1995, 61, 2771–2774. [Google Scholar] [CrossRef]
- Jepson, P.D.; Baker, J.R.; Kuiken, T.; Simpson, V.R.; Kennedy, S.; Bennett, P.M. Pulmonary pathology of harbour porpoises (Phocoena phocoena) stranded in England. Vet. Rec. 2000, 146, 721–728. [Google Scholar] [CrossRef]
- Dalebout, M.L.; Van Helden, A.; Van Waerebeek, K.; Baker, C.S. Molecular genetic identification of southern hemisphere beaked whales (Cetacea: Ziphiidae). Mol. Ecol. 1998, 7, 687–694. [Google Scholar] [CrossRef]
- Turner, S.; Pryer, K.M.; Miao, V.P.; Palmer, J.D. Investigating deep phylogenetic relationships among cyanobacteria and plastids by small subunit rRNA sequence analysis 1. J. Eukaryot. Microbiol. 1999, 46, 327–338. [Google Scholar] [CrossRef]
- Pu, Z.Y.; Dobos, M.; Limsowtin, G.K.Y.; Powell, I.B. Integrated polymerase chain reaction-based procedures for the detection and identification of species and subspecies of the Gram-positive bacterial genus Lactococcus. J. Appl. Microbiol. 2002, 93, 353–361. [Google Scholar] [CrossRef]
- Lane, D.J. 16S/23S rRNA sequencing. In Nucleic Acid Techniques in Bacterial Systematics; Stackebrandt, E., Goodfellow, M., Eds.; John Wiley & Sons: New York, NY, USA, 1991; pp. 115–175. [Google Scholar]
- Nomura, M.; Kobayashi, M.; Okamoto, T. Rapid PCR-based method which can determine both phenotype and genotype of Lactococcus lactis subspecies. Appl. Environ. Microbiol. 2002, 68, 2209–2213. [Google Scholar] [CrossRef]
- Yoon, S.H.; Ha, S.M.; Kwon, S.; Lim, J.; Kim, Y.; Seo, H.; Chun, J. Introducing EzBioCloud: A taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 2017, 67, 1613–1617. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [PubMed]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef] [PubMed]
- Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Nomura, M.; Kimoto, H.; Someya, Y.; Suzuki, I. Novel characteristic for distinguishing Lactococcus lactis subsp. lactis from subsp. cremoris. Int. J. Syst. Evol. Microbiol. 1999, 49, 163–166. [Google Scholar] [CrossRef]
- Huey, B.I.N.G.; Hall, J.E.F.F. Hypervariable DNA fingerprinting in Escherichia coli: Minisatellite probe from bacteriophage M13. J. Bacteriol. 1989, 171, 2528–2532. [Google Scholar] [CrossRef]
- Ryu, E. A simple method of differentiation between Gram-positive and Gram-negative organisms without staining. Kitasato Arch. Exp. Med. 1940, 17, 58–63. [Google Scholar]
- Neveling, D.P.; Ahire, J.J.; Laubscher, W.; Rautenbach, M.; Dicks, L.M. Genetic and phenotypic characteristics of a multi-strain probiotic for broilers. Curr. Microbiol. 2020, 77, 369–387. [Google Scholar] [CrossRef]
- Artimo, P.; Jonnalagedda, M.; Arnold, K.; Baratin, D.; Csardi, G.; De Castro, E.; Duvaud, S.; Flegel, V.; Fortier, A.; Gasteiger, E.; et al. ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 2012, 40, 597–603. [Google Scholar] [CrossRef]
- Agrawal, P.; Khater, S.; Gupta, M.; Sain, N.; Mohanty, D. RiPPMiner: A bioinformatics resource for deciphering chemical structures of RiPPs based on prediction of cleavage and cross-links. Nucleic Acids Res. 2017, 45, 80–88. [Google Scholar] [CrossRef] [PubMed]
- Alegría, Á.; Delgado, S.; Roces, C.; López, B.; Mayo, B. Bacteriocins produced by wild Lactococcus lactis strains isolated from traditional, starter-free cheeses made of raw milk. Int. J. Food Microbiol. 2010, 143, 61–66. [Google Scholar] [CrossRef] [PubMed]
- Martinez, B.; Suarez, J.E.; Rodriguez, A. Antimicrobials produced by wild lactococcal strains isolated from homemade cheeses. J. Food Prot. 1995, 58, 1118–1123. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Cuesta, M.C.; Kok, J.; Herranz, E.; Peláez, C.; Requena, T.; Buist, G. Requirement of autolytic activity for bacteriocin-induced lysis. Appl. Environ. Microbiol. 2000, 66, 3174–3179. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Martinez, B.; Fernandez, M.; Suarez, J.E.; Rodriguez, A. Synthesis of lactococcin 972, a bacteriocin produced by Lactococcus lactis IPLA 972, depends on the expression of a plasmid-encoded bicistronic operon. Microbiology 1999, 145, 3155–3162. [Google Scholar] [CrossRef][Green Version]
- Van Belkum, M.J.; Hayema, B.J.; Jeeninga, R.E.; Kok, J.; Venema, G. Organization and nucleotide sequences of two lactococcal bacteriocin operons. Appl. Environ. Microbiol. 1991, 57, 492–498. [Google Scholar] [CrossRef] [PubMed]
- Olasupo, N.A.; Schillinger, U.; Narbad, A.; Dodd, H.; Holzapfel, W.H. Occurrence of nisin Z production in Lactococcus lactis BFE 1500 isolated from wara, a traditional Nigerian cheese product. Int. J. Food Microbiol. 1999, 53, 141–152. [Google Scholar] [CrossRef]
- Li, H.; O’Sullivan, D.J. Heterologous expression of the Lactococcus lactis bacteriocin, nisin, in a dairy Enterococcus strain. Appl. Environ. Microbiol. 2002, 68, 3392–3400. [Google Scholar] [CrossRef][Green Version]
- Ghrairi, T.; Manai, M.; Berjeaud, J.M.; Frere, J. Antilisterial activity of lactic acid bacteria isolated from rigouta, a traditional Tunisian cheese. J. Appl. Microbiol. 2004, 97, 621–628. [Google Scholar] [CrossRef]
- Veljovic, K.; Terzic-Vidojevic, A.; Vukasinovic, M.; Strahinic, I.; Begovic, J.; Lozo, J.; Ostojic, M.; Topisirovic, L. Preliminary characterization of lactic acid bacteria isolated from Zlatar cheese. J. Appl. Microbiol. 2007, 103, 2142–2152. [Google Scholar] [CrossRef]
- Fusieger, A.; Perin, L.M.; Teixeira, C.G.; de Carvalho, A.F.; Nero, L.A. The ability of Lactococcus lactis subsp. lactis bv. diacetylactis strains in producing nisin. Antonie van Leeuwenhoek 2019, 113, 651–662. [Google Scholar] [CrossRef] [PubMed]
- Itoi, S.; Uchida, J.; Takanashi, S.; Narita, T.; Abe, K.; Naya, S.; Sugita, H. The clam Meretrix lamarckii (Bivalvia: Veneridae) is a rich repository of marine lactic acid bacterial strains. Ann. Microbiol. 2014, 64, 1267–1274. [Google Scholar] [CrossRef]
- Teuber, M.; Geis, A. The genus lactococcus. In The Prokaryotes, 3rd ed.; Dworkin, M., Falkow, S., Rosenberg, E., Schleifer, K.-H., Stackebrandt, E., Eds.; Springer: New York, NY, USA, 2006; Volume 4, pp. 205–228. [Google Scholar]
- Kimoto, H.; Nomura, M.; Kobayashi, M.; Okamoto, T.; Ohmomo, S. Identification and probiotic characteristics of Lactococcus strains from plant materials. Jpn. Agric. Res. Q. 2004, 38, 111–117. [Google Scholar] [CrossRef][Green Version]
- Klijn, N.; Weerkamp, A.H.; De Vos, W.M. Detection and characterization of lactose-utilizing Lactococcus spp. in natural ecosystems. Appl. Environ. Microbiol. 1995, 61, 788–792. [Google Scholar] [CrossRef] [PubMed]
- Shiina, A.; Itoi, S.; Washio, S.; Sugita, H. Molecular identification of intestinal microflora in Takifugu niphobles. Comp. Biochem. Physiol. Part D Genom. Proteom. 2006, 1, 128–132. [Google Scholar] [CrossRef]
- Itoi, S.; Yuasa, K.; Washio, S.; Abe, T.; Ikuno, E.; Sugita, H. Phenotypic variation in Lactococcus lactis subsp. lactis isolates derived from intestinal tracts of marine and freshwater fish. J. Appl. Microbiol. 2009, 107, 867–874. [Google Scholar] [CrossRef]
- Ohizumi, H.; Kishiro, T. Stomach contents of a Cuviers beaked whale (Ziphius cavirostris) stranded on the central Pacific coast of Japan. Aquat. Mamm. 2003, 29, 99–103. [Google Scholar] [CrossRef]
- Lubelski, J.; Rink, R.; Khusainov, R.; Moll, G.N.; Kuipers, O.P. Biosynthesis, immunity, regulation, mode of action and engineering of the model lantibiotic nisin. Cell. Mol. Life Sci. 2008, 65, 455–476. [Google Scholar] [CrossRef]
- Khusainov, R.; Heils, R.; Lubelski, J.; Moll, G.N.; Kuipers, O.P. Determining sites of interaction between prenisin and its modification enzymes NisB and NisC. Mol. Microbiol. 2011, 82, 706–718. [Google Scholar] [CrossRef]
- Qiao, M.; Saris, P.E. Evidence for a role of NisT in transport of the lantibiotic nisin produced by Lactococcus lactis N8. FEMS Microbiol. Lett. 1996, 144, 89–93. [Google Scholar] [CrossRef][Green Version]
- Lagedroste, M.; Smits, S.H.; Schmitt, L. Substrate specificity of the secreted nisin leader peptidase NisP. Biochemistry 2017, 56, 4005–4014. [Google Scholar] [CrossRef] [PubMed]
- Cheigh, C.I.; Pyun, Y.R. Nisin biosynthesis and its properties. Biotechnol. Lett. 2005, 27, 1641–1648. [Google Scholar] [CrossRef]
- Siegers, K.; Entian, K.D. Genes involved in immunity to the lantibiotic nisin produced by Lactococcus lactis 6F3. Appl. Environ. Microbiol. 1995, 61, 1082–1089. [Google Scholar] [CrossRef] [PubMed]
- AlKhatib, Z.; Lagedroste, M.; Zaschke, J.; Wagner, M.; Abts, A.; Fey, I.; Kleinschrodt, D.; Smits, S.H. The C-terminus of nisin is important for the ABC transporter Nis FEG to confer immunity in Lactococcus lactis. Microbiologyopen 2014, 3, 752–763. [Google Scholar] [CrossRef] [PubMed]
- Choi, H.J.; Cheigh, C.I.; Kim, S.B.; Pyun, Y.R. Production of a nisin-like bacteriocin by Lactococcus lactis subsp. lactis A164 isolated from Kimchi. J. Appl. Microbiol. 2000, 88, 563–571. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.L.; Kim, D.H. Genome-wide comparison reveals a probiotic strain Lactococcus lactis WFLU12 isolated from the gastrointestinal tract of olive flounder (Paralichthys olivaceus) harboring genes supporting probiotic action. Mar. Drugs 2018, 16, 140. [Google Scholar] [CrossRef] [PubMed]

| Use | Primer Name | Gene | Sequence (5′ to 3′) | Reference |
|---|---|---|---|---|
| Whale species identification | t-Pro whale | mitochondrial DNA D-loop region | TCACCCAAAGCTGRARTTCTA | [27] |
| Dlp5 | CCATCGWGATGTCTTATTTAAGRGGAA | |||
| Lactococcus lactis-specific PCR | 8F | 16S rRNA | AGAGTTTGATCCTGGCTCAG | [28] |
| LacreR | GGGATCATCTTTGAGTGAT | [29] | ||
| Bacterial identification and phylogenetic analysis | 27F | 16S rRNA | AGAGTTTGATCMTGGCTCAG | [30] |
| 1492R | TACGGYTACCTTGTTACGACTT | |||
| Classification of Lactococcus lactis subspecies | gadB21 | gadB | CGTTATGGATTTGATGGATATAAAGC | [31] |
| GAD7 | ACTCTTCTTAAGAACAAGTTTAACAGC |
| Antimicrobial Activity of the Cell-Free Supernatant (pH 7.0) | ||||||
|---|---|---|---|---|---|---|
| Indicator Bacteria Species | Strain | CBW1 | CBW2 | CBW3 | CBW4 | CBW5 |
| Vibrio alginolyticus | ATCC 17749 | − | ++ | − | − | − |
| Vibrio parahaemolyticus | ATCC 17802 | − | − | − | − | − |
| Escherichia coli | DSM 30083 | − | − | − | − | − |
| Photobacterium damselae subsp. damselae | DSM 7482 | − | − | − | − | − |
| Lactococcus lactis subsp. lactis | ATCC 19435 | − | − | − | − | − |
| Lactococcus lactis subsp. cremoris | ATCC 19257 | − | − | − | − | − |
| Lactococcus garvieae | ATCC 43921 | − | − | − | − | − |
| Lactococcus plantarum | ATCC 43199 | − | − | − | − | − |
| Lactococcus raffinolactis | ATCC 43920 | − | − | − | − | − |
| Enterococcus faecalis | DSM 20478 | − | − | − | − | − |
| Enterococcus hirae | ATCC 8043 | − | − | − | − | − |
| Enterococcus faecium | ATCC 19434 | − | − | − | − | − |
| Enterococcus canis | DSM 17029 | − | − | − | − | − |
| Staphylococcus xylosus | ATCC 29971 | − | − | − | − | − |
| Staphylococcus epidermidis | ATCC 14990 | − | − | − | − | − |
| Bacillus subtilis subsp. subtilis | ATCC 6051 | − | + | − | − | − |
| Streptococcus salivarius | DSM 20560 | − | − | − | − | − |
| Primer Name | Gene | Sequence (5′ to 3′) | Reference |
|---|---|---|---|
| Nis-F | nisA | CGGCTCTGATTAAATTCTGAAG | [43] |
| Nis-R | GGATTAGCTAGTAGTAACTGTTC | ||
| Lact481-F | Lacticin 481 | TCTGCACTCACTTCATTAGTTA | [44] |
| Lact482-R | AAGGTAATTACACCTCTTTTAT | ||
| Lact3147-F | Lacticin 3147 | TACTGGGGAAATAACGG | [45] |
| Lact3148-R | TGGACAAGTATTGGTAC | ||
| Lcn972-F | Lactococcin 972 | TTGTAGCTCCTGCAGAAGGAACATGG | [46] |
| Lcn973-R | GCCTTAGCTTTGAATTCTTACCAAAAG | ||
| LactABM-F | Lactococcin A, B, and M | GAAGAGGCAATCAGTAGAG | [43,47] |
| LactA-R | Lactococcin A gene | GTGTTCTATTTATAGCTAATG | [43] |
| LactB-R | Lactococcin B gene | CCAGGATTTTCTTTGATTTACTTC | [43] |
| LactM-R | Lactococcin M gene | GTGTACTGGTCTAGCATAAG | [47] |
| p4 | nisB | AGAGAAGTTATTTACGATCAAC | [48] |
| P5 | ATCTGACAACAAATCTTTTTGT | ||
| p6 | nisC | TTCAGAGCAATATGAGG | [48] |
| p7 | TATTAAGGCCACAATAAG | ||
| P06-F | nisT | GAAGAATACATGAAATGAGG | [49] |
| P06-R | TAACTTTCCAGCTGTCCC | ||
| P08-F | nisI | ATTGTGGCCTTAATAGGG | [49] |
| P08-R | TAGCGACTTGTCAGAAGC | ||
| P11-F | nisF | CAGGTGCTACAAGATATCAG | [49] |
| P11-R | ACAACTCCGCAATACCATCAG | ||
| Prim-NisP5 | nisP | GGATTTGGTATCTGTTTCGAAG | [50] |
| Prim-NisP3 | TCTTTCCCATTAACTTGTACTGTG | ||
| Nis3 | nisRK | CAGTGCCATGGGTAAAAAATATTCAATGCG | [51] |
| Nis4 | CTTAGAGAATTCTCTAATGAG |
| Strain | Sequence Length (bp) | Nearest Phylogenetic Neighbor | Similarity (%) | Completeness (%) |
|---|---|---|---|---|
| CBW1 | 1420 | Lactococcus lactis subsp. lactis JCM 5805T | 99.93 | 96.3 |
| CBW2 | 1414 | 99.65 | 95.9 | |
| CBW3 | 1412 | 99.86 | 95.8 | |
| CBW4 | 1368 | 99.78 | 92.2 | |
| CBW5 | 1376 | 99.04 | 91.8 |
| Characteristics | CBW1 | CBW2 | CBW3 | CBW4 | CBW5 |
|---|---|---|---|---|---|
| Gram reaction | + | + | + | + | + |
| Motility | − | − | − | − | − |
| Catalase | − | − | − | − | − |
| Gas from glucose | − | − | − | − | − |
| Final pH b | 4.25 | 4.31 | 4.32 | 4.29 | 4.34 |
| Growth | |||||
| NaCl (%) | |||||
| 1 | + | + | + | + | + |
| 2 | + | + | + | + | + |
| 3 | + | + | + | + | + |
| 4 | + | + | + | + | + |
| 5 | + | + | + | + | + |
| 6 | + | + | + | + | + |
| 7 | + | + | + | + | + |
| 8 | − | − | − | − | − |
| 9 | − | − | − | − | − |
| 10 | − | − | − | − | − |
| 15 | − | − | − | − | − |
| Temperature (°C) | |||||
| 10 | + | + | + | + | + |
| 40 | + | + | + | + | + |
| 45 | − | − | − | − | − |
| pH | |||||
| 4.0 | + | + | + | + | + |
| 9.2 | + | + | + | + | + |
| 10.0 | + | + | + | + | + |
| Diameter of the Inhibition Zone ± SD (mm) | ||
|---|---|---|
| Enzyme | Vibrio alginolyticus | Bacillus subtilis subsp. subtilis |
| Control | 15.13 ± 0.15 | 8.38 ± 0.34 |
| Proteinase K | 0.00 ± 0.00 ** | 0.00 ± 0.00 ** |
| Trypsin | 8.80 ± 0.20 ** | 7.86 ± 0.10 * |
| Pepsin | 10.13 ± 0.12 ** | 7.87 ± 0.25 * |
| Lysozyme | 15.13 ± 0.31 N.S. | 8.23 ± 0.16 N.S. |
| Catalase | 14.97 ± 0.25 N.S. | 8.30 ± 0.20 N.S. |
| Lipase | 14.83 ± 0.15 N.S. | 8.26 ± 0.25 N.S. |
| Gene | CBW1 | CBW2 | CBW3 | CBW4 | CBW5 |
|---|---|---|---|---|---|
| nisA | − | + | − | − | − |
| nisB | − | + | − | − | − |
| nisC | − | + | − | − | − |
| nisT | − | + | − | − | − |
| nisI | − | + | − | − | − |
| nisF | − | + | − | − | − |
| nisP | − | + | − | − | − |
| nisRK | − | + | − | − | − |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Suzuki, A.; Suzuki, M. Antimicrobial Activity of Lactococcus lactis subsp. lactis Isolated from a Stranded Cuvier’s Beaked Whale (Ziphius cavirostris) against Gram-Positive and -Negative Bacteria. Microorganisms 2021, 9, 243. https://doi.org/10.3390/microorganisms9020243
Suzuki A, Suzuki M. Antimicrobial Activity of Lactococcus lactis subsp. lactis Isolated from a Stranded Cuvier’s Beaked Whale (Ziphius cavirostris) against Gram-Positive and -Negative Bacteria. Microorganisms. 2021; 9(2):243. https://doi.org/10.3390/microorganisms9020243
Chicago/Turabian StyleSuzuki, Akihiko, and Miwa Suzuki. 2021. "Antimicrobial Activity of Lactococcus lactis subsp. lactis Isolated from a Stranded Cuvier’s Beaked Whale (Ziphius cavirostris) against Gram-Positive and -Negative Bacteria" Microorganisms 9, no. 2: 243. https://doi.org/10.3390/microorganisms9020243
APA StyleSuzuki, A., & Suzuki, M. (2021). Antimicrobial Activity of Lactococcus lactis subsp. lactis Isolated from a Stranded Cuvier’s Beaked Whale (Ziphius cavirostris) against Gram-Positive and -Negative Bacteria. Microorganisms, 9(2), 243. https://doi.org/10.3390/microorganisms9020243






