Bacterial Diversity and Community Composition Distribution in Cold-Desert Habitats of Qinghai–Tibet Plateau, China
Abstract
:1. Introduction
2. Materials and Methods
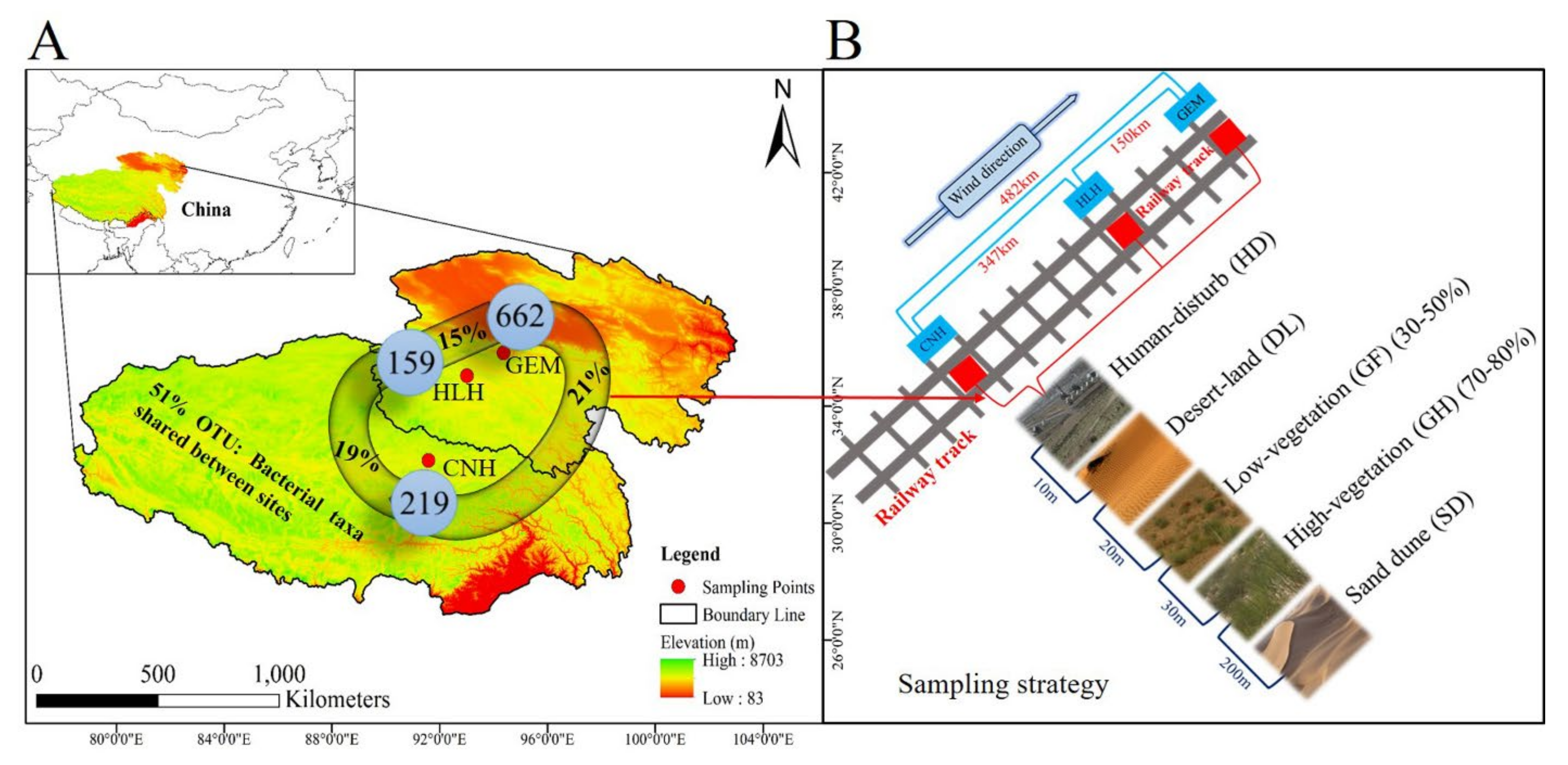
2.1. Study Site Description
2.2. Experimental Design and Sampling Procedure
2.3. Soil Physicochemical Analysis
2.4. DNA Extraction, PCR Amplification, and Illumina MiSeq Sequencing
2.5. Bioinformatics Analysis
2.6. Statistical Analysis and Data Visualization
3. Results
3.1. Soil Characteristics of the Sampling Sites
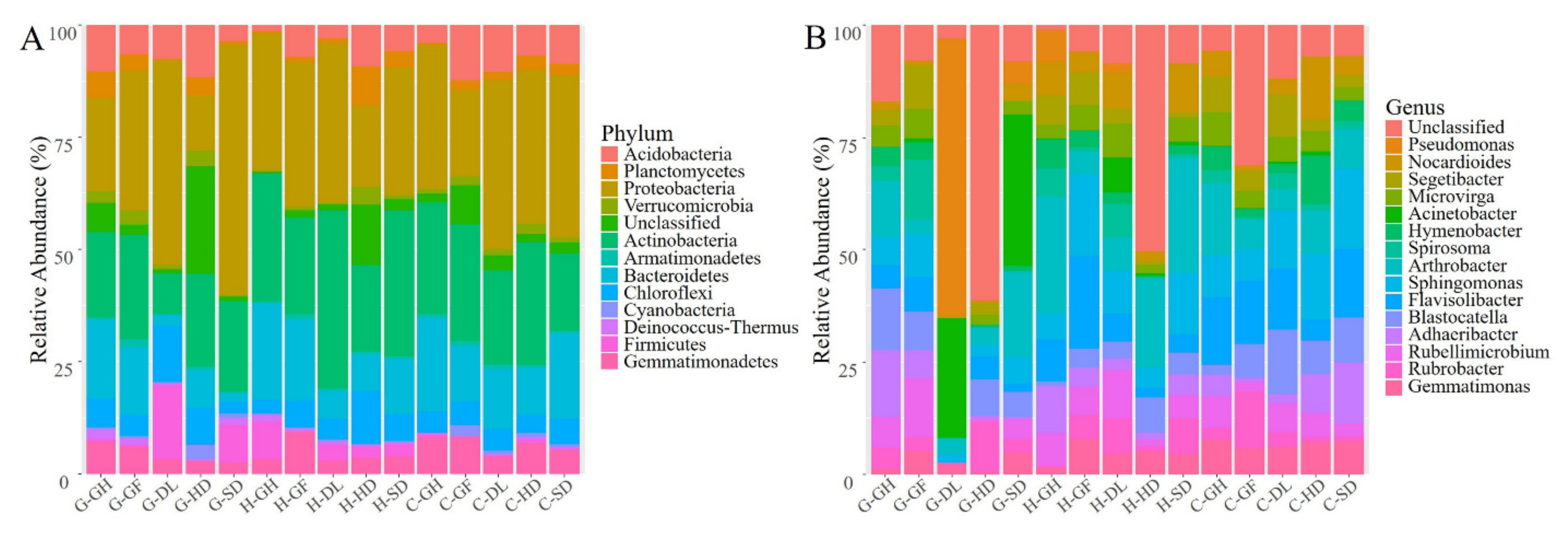
3.2. Bacterial Richness, Diversity, and Taxonomic Composition
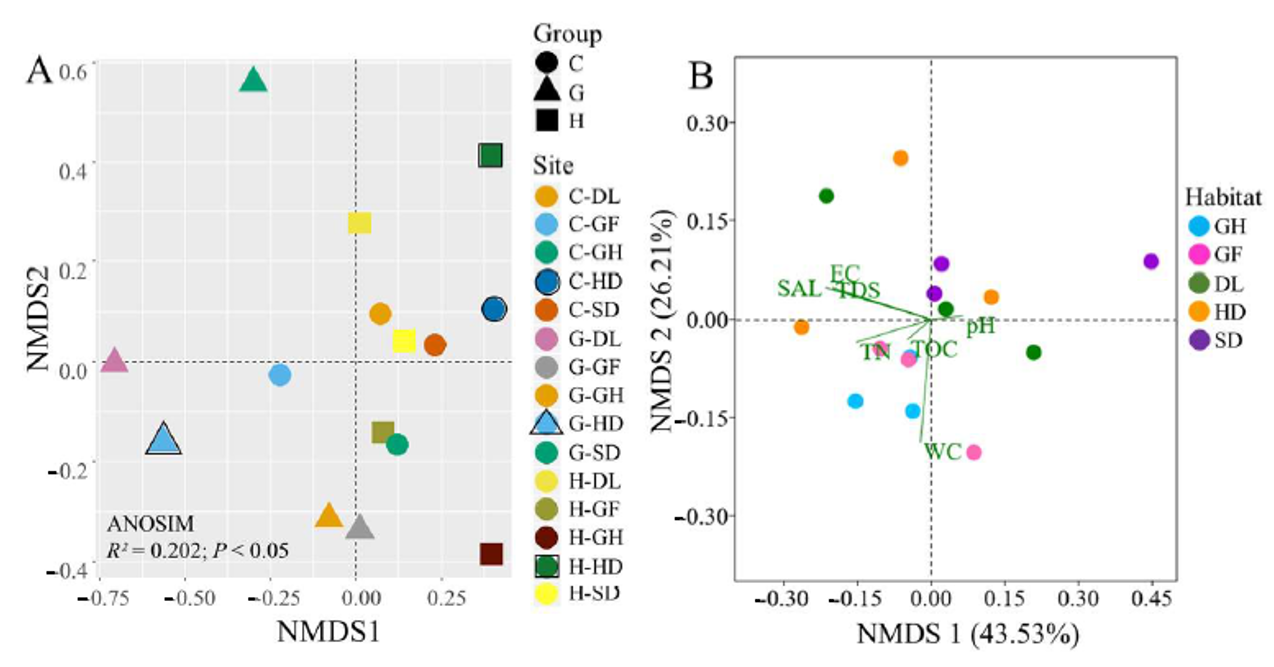
3.3. Distribution of Bacterial Community Across the Habitats
3.4. Identification of Environmental Variables Shaping Cold-Desert Bacterial Community Structures
3.5. Indicator Taxa in Soil Bacterial Communities
4. Discussion
4.1. Variation in Bacterial Diversity and Community Composition
4.2. Bacterial Communities in Relation to Environmental Variables
4.3. Network Interaction Differences and Potential Indicators
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zeng, B.; Yang, T.B. Natural vegetation responses to warming climates in Qaidam Basin 1982–2003. Int. J. Remote Sens. 2009, 30, 5685–5701. [Google Scholar] [CrossRef]
- Li, M.; Fang, X.; Yi, C.; Gao, S.; Zhang, W.; Galy, A. Evaporite minerals and geochemistry of the upper 400 m sediments in a core from the Western Qaidam Basin, Tibet. Quat. Int. 2010, 218, 176–189. [Google Scholar] [CrossRef]
- Sul, W.J.; Asuming-Brempong, S.; Wang, Q.; Tourlousse, D.M.; Penton, C.R.; Deng, J.L.M. Tropical agricultural land management influences on soil microbial communities through its effect on soil organic carbon. Soil Biol. Biochem. 2013, 65, 33–38. [Google Scholar] [CrossRef] [Green Version]
- Xing, R.; Gao, Q.B.; Zhang, F.Q.; Wang, J.L.; Chen, S.L. Large-scale distribution of bacterial communities in the Qaidam Basin of the Qinghai-Tibet Plateau. Microbiologyopen 2019, 8, e909. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Bahadur, A.; Zhang, G.; Zhang, B.; Wu, X.; Chen, T.; Liu, G. Diverse bacterial communities from Qaidam Basin of the Qinghai-Tibet Plateau: Insights into variations in bacterial diversity across different regions. Front. Microbiol. 2020, 11, 554105. [Google Scholar] [CrossRef] [PubMed]
- Makhalanyane, T.P.; Valverde, A.; Gunnigle, E.; Frossard, A.; Ramond, J.B.; Cowan, D.A. Microbial ecology of hot desert edaphic systems. FEMS Microbiol. Rev. 2015, 39, 203–221. [Google Scholar] [CrossRef]
- Whitman, W.B.; Coleman, D.C.; Wiebe, W.J. Prokaryotes: The unseen majority. Proc. Natl. Acad. Sci. USA 1998, 95, 6578–6583. [Google Scholar] [CrossRef] [Green Version]
- Venter, J.C.; Remington, K.; Heidelberg, J.F.; Halpern, A.L.; Rusch, D.; Eisen, J.A.; Wu, D.; Paulsen, I.; Nelson, K.E.; Nelson, W.; et al. Environmental genome shotgun sequencing of the Sargasso Sea. Science 2004, 304, 66–74. [Google Scholar] [CrossRef] [Green Version]
- Shen, S. Community structure and diversity of culturable moderate halophilic bacteria isolated from Qrhan Salt Lake on Qinghai-Tibet Plateau. Acta Ecol. Sin. 2017, 57, 490–499. [Google Scholar]
- Gallardo, A.; Schlesinger, W.H. Carbon and nitrogen limitations of soil microbial biomass in desert ecosystems. Biogeochemistry 1992, 18, 1–17. [Google Scholar] [CrossRef]
- Tin, T.; Fleming, Z.L.; Hughes, K.A.; Ainley, D.G.; Convey, P.; Moreno, C.A.; Pfeiffer, S.; Scott, J.; Snape, I. Impacts of local human activities on the Antarctic environment, Antarctic. Science 2008, 21, 3–33. [Google Scholar] [CrossRef] [Green Version]
- Cederlund, H.; Stenstrom, J. Microbial biomass and activity on railway track and embankments. Pest Manag. Sci. 2004, 60, 550–555. [Google Scholar] [CrossRef] [PubMed]
- Griffiths, R.I.; Thomson, B.C.; James, P.; Bell, T.; Bailey, M.; Whiteley, A.S. The bacterial biogeography of British soils. Environ. Microbiol. 2011, 13, 1642–1654. [Google Scholar] [CrossRef]
- Wang, J.; Zhang, T.; Li, L.; Li, J.; Feng, Y.; Lu, Q. The patterns and drivers of bacterial and fungal beta-diversity in a typical dryland ecosystem of northwest China. Front. Microbiol. 2017, 8, 2126. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Whitaker, R.J.; Grogan, D.W.; Taylor, J.W. Geographic barriers isolate endemic populations of hyperthermophilic archaea. Science 2003, 301, 976–978. [Google Scholar] [CrossRef]
- Green, J.L.; Bohannan, B.J.M.; Whitaker, R.J. Microbial biogeography: From taxonomy to traits. Science 2008, 320, 1039–1043. [Google Scholar] [CrossRef] [Green Version]
- Fierer, N.; Jackson, R.B. The diversity and biogeography of soil bacterial communities. Proc. Natl. Acad. Sci. USA 2006, 103, 626–631. [Google Scholar] [CrossRef] [Green Version]
- Stegen, J.C.; Lin, X.; Fredrickson, J.K.; Chen, X.; Kennedy, D.W.; Murray, C.J.; Rockhold, M.L.; Konopka, A. Quantifying community assembly processes and identifying features that impose them. Int. Soc. Microb. Ecol. J. 2013, 7, 2069–2079. [Google Scholar] [CrossRef]
- Tripathi, B.M.; Stegen, J.C.; Kim, M.; Dong, K.; Adams, J.M.; Lee, Y.K. Soil pH mediates the balance between stochastic and deterministic assembly of bacteria. Int. Soc. Microb. Ecol. J. 2018, 12, 1072–1083. [Google Scholar] [CrossRef]
- Zhang, B.; Wu, X.; Tai, X.; Sun, L.; Wu, M.; Zhang, W.; Chen, X.; Zhang, G.; Chen, T.; Liu, G.; et al. Variation in actinobacterial community composition and potential function in different soil ecosystems belonging to the arid Heihe River Basin of northwest China. Front. Microbiol. 2019, 10, 2209. [Google Scholar] [CrossRef]
- Miller, S.R.; Strong, A.L.; Jones, K.L.; Ungerer, M.C. Bar-coded pyrosequencing reveals shared bacterial community properties along the temperature gradients of two alkaline hot springs in Yellowstone National Park. Appl. Environ. Microbiol. 2009, 75, 4565–4572. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hansel, C.M.; Fendorf, S.; Jardine, P.M.; Francis, C.A. Changes in bacterial and archaeal community structure and functional diversity along a geochemically variable soil profile. Appl. Environ. Microbiol. 2008, 74, 1620–1633. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crits-Christoph, A.; Robinson, C.K.; Barnum, T.; Fricke, W.F.; Davila, A.F.; Jedynak, B.; McKay, C.P.; DiRuggiero, J. Colonization patterns of soil microbial communities in the Atacama Desert. Microbiome 2013, 1, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Valverde, A.; Makhalanyane, T.P.; Seely, M.; Cowan, D.A. Cyanobacteria drive community composition and functionality in rock-soil interface communities. Mol. Ecol. 2015, 24, 812–821. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, A.C.; Stubbs, T. Soil microbial communities as indicators of soil health. Ann. Arid Zone 2006, 45, 1–13. [Google Scholar]
- Fan, M.; Li, J.; Tang, Z.; Shangguan, Z. Soil bacterial community succession during desertification in a desert steppe ecosystem. Land Degrad. Dev. 2020, 31, 1662–1674. [Google Scholar] [CrossRef]
- Liebner, S.; Rublack, K.; Stuehrmann, T.; Wagner, D. Diversity of aerobic methanotrophic bacteria in a permafrost active layer soil of the Lena Delta, Siberia. Microb. Ecol. 2009, 57, 25–35. [Google Scholar] [CrossRef] [Green Version]
- Zhang, B.; Wu, X.; Zhang, G.; Li, S.; Zhang, W.; Chen, X. The diversity and biogeography of the communities of actinobacteria in the forelands of glaciers at a continental scale. Environ. Res. Lett. 2016, 11, 054012. [Google Scholar] [CrossRef]
- Wickham, H.; Chang, W. ggplot2 An Implementation of the Grammar of Graphics. 2015. Available online: https://www.r-project.org/conferences/useR-2006/Abstracts/Wickham.pdf (accessed on 5 October 2020).
- Chao, A. Non-parametric estimation of the classes in a population. Scand. J. Stat. 1984, 11, 265–270. [Google Scholar]
- Oksanen, J.; Blanchet, F.G.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.E.A. Vegan: Community Ecology Package. 2017. Available online: https://CRAN.Rproject.org/package=vegan (accessed on 5 October 2020).
- Clarke, K.R.; Warwick, R.M. Similarity-based testing for community pattern: The two-way layout with no replication. Mar. Biol. 1994, 118, 167–176. [Google Scholar] [CrossRef]
- Henderson-Sellers, A.; Irannejad, P.; McGuffie, K. Future desertification and climate change: The need for land-surface system evaluation improvement. Glob. Planet. Chang. 2008, 64, 129–138. [Google Scholar] [CrossRef]
- Chen, H.; Boutros, P.C. VennDiagram: A package for the generation of highly-customizable Venn and Euler diagrams in R. BMC Bioinform. 2011, 12, 35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Griffiths, R.I.; Thomson, B.C.; Plassart, P.; Gweon, H.S.; Stone, D.; Creamer, R.E.; Lemanceau, P.; Bailey, M.J. Mapping and validating predictions of soil bacterial biodiversity using European and national scale datasets. Appl. Soil Ecol. 2016, 97, 61–68. [Google Scholar] [CrossRef] [Green Version]
- Sajjad, W.; Din, G.; Rafiq, M.; Iqbal, A.; Khan, S.; Zada, S.; Ali, B.; Kang, S. Pigment production by cold-adapted bacteria and fungi: Colorful tale of cryosphere with wide range applications. Extremophiles 2020, 24, 447–473. [Google Scholar] [CrossRef] [PubMed]
- Han, Q.; Huang, J.; Long, D.; Wang, X.; Liu, J. Diversity and community structure of ectomycorrhizal fungi associated with Larix chinensis across the alpine treeline ecotone of Taibai Mountain. Mycorrhiza 2017, 27, 487–497. [Google Scholar] [CrossRef]
- Wang, W.; Yujun, Y.; Yang, Y.; Zhou, Y.; Zhang, S.; Wang, X.; Yang, Z. Impact of anthropogenic activities on the sediment microbial communities of Baiyangdian shallow lake. Int. J. Sediment Res. 2020, 35, 180–192. [Google Scholar] [CrossRef]
- Tang, J.; Tang, X.; Qin, Y.; He, Q.; Yi, Y.; Ji, Z. Karst rocky desertification progress: Soil calcium as a possible driving force. Sci. Total Environ. 2019, 649, 1250–1259. [Google Scholar] [CrossRef]
- Mapelli, F.; Marasco, R.; Fusi, M.; Scaglia, B.; Tsiamis, G.; Rolli, E.; Fodelianakis, S.; Bourtzis, K.; Ventura, S.; Tambone, F.; et al. The stage of soil development modulates rhizosphere effect along a High Arctic desert chronosequence. Int. Soc. Microb. Ecol. J. 2018, 12, 1188–1198. [Google Scholar] [CrossRef]
- Sun, Y.; Shi, Y.L.; Wang, H.; Zhang, T.; Yu, L.Y.; Sun, H.; Zhang, Y.Q. Diversity of bacteria and the characteristics of actinobacteria community structure in Badain Jaran desert and Tengger desert of China. Front. Microbiol. 2018, 9, 1068. [Google Scholar] [CrossRef] [Green Version]
- Van Horn, D.J.; Van Horn, M.L.; Barrett, J.E.; Gooseff, M.N.; Altrichter, A.E.; Geyer, K.M.; Zeglin, L.H.; Takacs-Vesbach, C.D. Factors controlling soil microbial biomass and bacterial diversity and community composition in a cold desert ecosystem: Role of geographic scale. PLoS ONE 2013, 8, e66103. [Google Scholar] [CrossRef]
- Quoreshi, A.M.; Suleiman, M.K.; Kumar, V.; Manuvel, A.J.; Sivadasan, M.T.; Islam, M.A.; Khasa, D.P. Untangling the bacterial community composition and structure in selected Kuwait desert soils. Appl. Soil Ecol. 2019, 138, 1–9. [Google Scholar] [CrossRef]
- Gupta, P.; Sangwan, N.; Lal, R.; Vakhlu, J. Bacterial diversity of Drass, cold desert in Western Himalaya, and its comparison with Antarctic and Arctic. Arch. Microbiol. 2015, 197, 851–860. [Google Scholar] [CrossRef] [PubMed]
- Youssef, N.H.; Couger, M.B.; Elshahed, M.S. Fine-scale bacterial beta diversity within a complex ecosystem (Zodletone Spring, OK, USA): The role of the rare biosphere. PLoS ONE 2010, 5, e12414. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- An, S.; Couteau, C.; Luo, F.; Neveu, J.; DuBow, M.S. Bacterial diversity of surface sand samples from the Gobi and Taklamaken deserts. Microb. Ecol. 2013, 66, 850–860. [Google Scholar] [CrossRef]
- Costello, E.K.; Schmidt, S.K. Microbial diversity in alpine tundra wet meadow soil: Novel Chloroflexi from a cold, water-saturated environment. Environ. Microbiol. 2006, 8, 1471–1486. [Google Scholar] [CrossRef]
- Wagner, D.; Kobabe, S.; Liebner, S. Bacterial community structure and carbon turnover in permafrost-affected soils of the Lena Delta, northeastern Siberia. Can. J. Microbiol. 2009, 55, 73–83. [Google Scholar] [CrossRef] [Green Version]
- Pradhan, S.; Srinivas, T.N.R.; Pindi, P.K.; Kishore, K.H.; Begum, Z.; Singh, P.K.; Singh, A.K.; Pratibha, M.S.; Yasala, A.K.; Reddy, G.S.N.; et al. Bacterial biodiversity from Roopkund Glacier, Himalayan mountain ranges, India. Extremophiles 2010, 14, 377–395. [Google Scholar] [CrossRef]
- Shivaji, S.; Pratibha, M.S.; Sailaja, B.; Kishore, K.H.; Singh, A.K.; Begum, Z.; Anarasi, U.; Prabagaran, S.R.; Reddy, G.S.N.; Srinivas, T.N.R. Bacterial diversity of soil in the vicinity of Pindari glacier, Himalayan mountain ranges, India, using culturable bacteria and soil 16S rRNA gene clones. Extremophiles 2011, 15, 1–22. [Google Scholar] [CrossRef]
- Scola, V.; Ramond, J.B.; Frossard, A.; Zablocki, O.; Adriaenssens, E.M.; Johnson, R.M.; Seely, M.; Cowan, D.A. Namib desert soil microbial community diversity, assembly, and function along a natural Xeric Gradient. Microb. Ecol. 2018, 75, 193–203. [Google Scholar] [CrossRef] [Green Version]
- Gao, S.; Liang, J.; Teng, T.M. Petroleum contamination evaluation and bacterial community distribution in a historic oilfield located in loess plateau in China. Appl. Soil Ecol. 2019, 136, 30–42. [Google Scholar] [CrossRef]
- Liu, L.; Liu, Y.; Hui, R.; Xie, M. Recovery of microbial community structure of biological soil crusts in successional stages of Shapotou desert revegetation, northwest China. Soil Biol. Biochem. 2017, 107, 125–128. [Google Scholar] [CrossRef]
- Takahashi, Y.; Matsumoto, A.; Seino, A.; Iwai, Y.; Omura, S. Rare actinomycetes isolated from desert soils. Actinomycetologica 1996, 10, 91–97. [Google Scholar] [CrossRef]
- Sajjad, W.; Zheng, G.; Zhang, G.; Ma, X.; Xu, W.; Ali, B.; Rafiq, M. Diversity of prokaryotic communities indigenous to acid mine drainage and related rocks from Baiyin Open-Pit Copper Mine Stope, China. Geomicrobiol. J. 2018, 35, 580–600. [Google Scholar] [CrossRef]
- Stomeo, F.; Makhalanyane, T.P.; Valverde, A.; Pointing, S.B.; Stevens, M.I.; Cary, C.S.; Tuffin, M.I.; Cowan, D.A. Abiotic factors influence microbial diversity in permanently cold soil horizons of a maritime-associated Antarctic Dry Valley. FEMS Microbiol. Ecol. 2012, 82, 326–340. [Google Scholar] [CrossRef]
- Sher, Y.; Zaady, E.; Nejidat, A. Spatial and temporal diversity and abundance of ammonia oxidizers in semi-arid and arid soils: Indications for a differential seasonal effect on archaeal and bacterial ammonia oxidizers. FEMS Microbiol. Ecol. 2013, 86, 544–556. [Google Scholar] [CrossRef] [PubMed]
- Andrew, D.R.; Fitak, R.R.; Munguia-Vega, A.; Racolta, A.; Martinson, V.G.; Dontsova, K. Abiotic factors shape microbial diversity in Sonoran Desert soils. Appl. Environ. Microbiol. 2012, 78, 7527–7537. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Van Nostrand, J.D.; Deng, Y.; Lu, X.; Wang, C.; Zhou, J.; Han, X. Scale-dependent effects of climate and geographic distance on bacterial diversity patterns across northern China’s grasslands. FEMS Microbiol. Ecol. 2015, 91, fiv133. [Google Scholar] [CrossRef] [Green Version]
- Ganzert, L.; Lipski, A.; Hubberten, H.W.; Wagner, D. The impact of different soil parameters on the community structure of dominant bacteria from nine different soils located on Livingston Island, South Shetland Archipelago, Antarctica. FEMS Microbiol. Ecol. 2011, 76, 476–491. [Google Scholar] [CrossRef] [Green Version]
- Arocha-Garza, H.F.; Canales-Del Castillo, R.; Eguiarte, L.E.; Souza, V.; De la Torre-Zavala, S. High diversity and suggested endemicity of culturable Actinobacteria in an extremely oligotrophic desert oasis. PeerJ 2017, 5, e3247. [Google Scholar] [CrossRef] [Green Version]
- Varin, T.; Lovejoy, C.; Jungblut, A.D.; Vincent, W.F.; Corbeil, J. Metagenomic analysis of stress genes in microbial mat communities from Antarctica and the High Arctic. Appl. Environ. Microbiol. 2012, 78, 549–559. [Google Scholar] [CrossRef] [Green Version]
- Inskeep, W.P.; Jay, Z.J.; Tringe, S.G.; Herrgard, M.J.; Rusch, D.B.; Committee, M. Working Group, The YNP Metagenome project: Environmental parameters responsible for microbial distribution in the yellowstone geothermal ecosystem. Front. Microbiol. 2013, 4, 67. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tecon, R.; Or, D. Biophysical processes supporting the diversity of microbial life in soil. FEMS Microbiol. Rev. 2017, 41, 599–623. [Google Scholar] [CrossRef] [PubMed]
- Chu, H.; Sun, H.; Tripathi, B.M.; Adams, J.M.; Huang, R.; Zhang, Y.; Shi, Y. Bacterial community dissimilarity between the surface and subsurface soils equals horizontal differences over several kilometers in the western Tibetan Plateau. Environ. Microbiol. 2016, 18, 1523–1533. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Q.; Yang, X. Sedimentological and geochemical composition of aeolian sediments in the Taklamakan desert: Implications for provenance and sediment supply mechanisms. J. Geophys. Res. 2019, 124, 1217–1237. [Google Scholar] [CrossRef]
- Ronca, S.; Ramond, J.B.; Jones, B.E.; Seely, M.; Cowan, D.A. Namib Desert dune/interdune transects exhibit habitat-specific edaphic bacterial communities. Front. Microbiol. 2015, 6, 845. [Google Scholar] [CrossRef]
- Waheed, H.; Hashmi, I.; Naveed, A.K.; Khan, S.J. Molecular detection of microbial community in a nitrifying–denitrifying activated sludge system. Int. Biodeterior. Biodegrad. 2013, 85, 527–532. [Google Scholar] [CrossRef]
- Li, J.; Liu, W.; Luo, L.; Dong, D.; Liu, T.; Zhang, T.; Lu, C.; Liu, D.; Zhang, D.; Wu, H. Expression of Paenibacillus polymyxa β-1,3-1,4-glucanase in Streptomyces lydicus A01 improves its biocontrol effect against Botrytis cinerea. Biolog. Control. 2015, 90, 141–147. [Google Scholar] [CrossRef] [Green Version]
- Fierer, N.; Leff, J.W.; Adams, B.J.; Nielsen, U.N.; Bates, S.T.; Lauber, C.L.; Owens, S.; Gilbert, J.A.; Wall, D.H.; Caporaso, J.G. Cross-biome metagenomic analyses of soil microbial communities and their functional attributes. Proc. Natl. Acad. Sci. USA 2012, 109, 21390–21395. [Google Scholar] [CrossRef] [Green Version]
- De Cáceres, M.; Legendre, P.; Moretti, M. Improving indicator species analysis by combining groups of sites. Oikos 2010, 119, 1674–1684. [Google Scholar] [CrossRef]
- Tapia-Torres, Y.; López-Lozano, N.E.; Souza, V.; García-Oliva, F. Vegetation-soil system controls soil mechanisms for nitrogen transformations in an oligotrophic Mexican desert. J. Arid Environ. 2015, 114, 62–69. [Google Scholar] [CrossRef]
- Zhang, B.; Kong, W.; Wu, N.; Zhang, Y. Bacterial diversity and community along the succession of biological soil crusts in the Gurbantunggut Desert, Northern China. J. Basic Microbiol. 2016, 56, 670–679. [Google Scholar] [CrossRef] [PubMed]
- Long, H.; Kucukyildirim, S.; Sung, W.; Williams, E.; Lee, H.; Ackerman, M.; Doak, T.G.; Tang, H.; Lynch, M. Background mutational features of the radiation-resistant bacterium Deinococcus radiodurans. Mol. Biol. Evol. 2015, 32, 2383–2392. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sajjad, W.; Ali, B.; Bahadur, A.; Ghimire, P.S.; Kang, S. Bacterial diversity and communities structural dynamics in soil and meltwater runoff at the frontier of Baishui Glacier No.1, China. Microb. Ecol. 2020. [Google Scholar] [CrossRef] [PubMed]




| Site | pH | EC (μS/cm) | TDS (mg/L) | SAL (psu) | WC (%) | TN (%) | TOC (%) |
|---|---|---|---|---|---|---|---|
| G-GH | 7.88 ± 0.03a | 61.82 ± 1.50c | 31.19 ± 0.46c | 0.04 ± 0.01b | 9.18 ± 1.04b | 0.04 ± 0.00a | 1.05 ± 0.13b |
| G-GF | 7.81 ± 0.09a | 46.18 ± 0.69b | 23.31 ± 0.46b | 0.03 ± 0.00b | 6.74 ± 1.05ab | 0.03 ± 0.00a | 0.38 ± 0.02a |
| G-DL | 7.92 ± 0.07a | 45.42 ± 0.25b | 22.68 ± 0.07b | 0.03 ± 0.00b | 5.58 ± 0.40ab | 0.04 ± 0.00a | 0.49 ± 0.02a |
| G-HD | 8.29 ± 0.28a | 89.63 ± 0.76d | 45.12 ± 0.27d | 0.06 ± 0.01c | 4.42 ± 0.86a | 0.06 ± 0.00b | 0.50 ± 0.01a |
| G-SD | 7.84 ± 0.01a | 36.31 ± 0.88a | 17.79 ± 0.66a | 0.02 ± 0.01a | 4.87 ± 0.32ab | 0.03 ± 0.00a | 0.48 ± 0.08a |
| F | 2.00 | 534.45 | 616.12 | 13.83 | 5.92 | 17.28 | 14.71 |
| p | >0.05 | 0.000 | 0.000 | 0.007 | 0.039 | 0.004 | 0.006 |
| H-GH | 7.62 ± 0.11a | 33.29 ± 0.86b | 16.73 ± 0.41b | 0.02 ± 0.00a | 10.65 ± 0.50c | 0.04 ± 0.00a | 0.38 ± 0.04a |
| H-GF | 8.33 ± 0.32a | 37.31 ± 0.88b | 17.92 ± 0.79b | 0.03 ± 0.00ab | 5.18 ± 0.89b | 0.04 ± 0.01a | 0.36 ± 0.01a |
| H-DL | 8.30 ± 0.25a | 26.42 ± 0.86a | 13.20 ± 0.55a | 0.02 ± 0.00a | 10.28 ± 0.45c | 0.04 ± 0.01a | 0.31 ± 0.06a |
| H-HD | 8.57 ± 0.39a | 64.87 ± 0.90d | 32.42 ± 0.30d | 0.04 ± 0.01b | 4.29 ± 1.29ab | 0.04 ± 0.00a | 0.39 ± 0.04a |
| H-SD | 7.98 ± 0.01a | 44.15 ± 1.83c | 22.84 ± 1.08c | 0.04 ± 0.01ab | 1.86 ± 0.65a | 0.04 ± 0.00a | 0.34 ± 0.00a |
| F | 1.97 | 168.88 | 118.24 | 3.20 | 22.35 | 0.53 | 0.71 |
| p | >0.05 | 0.000 | 0.000 | 0.177 | 0.002 | 0.720 | 0.62 |
| C-GH | 7.73 ± 0.07a | 45.71 ± 2.17ab | 23.32 ± 0.78ab | 0.03 ± 0.01a | 11.13 ± 2.39c | 0.05 ± 0.00a | 0.72 ± 0.03a |
| C-GF | 7.76 ± 0.01ab | 52.45 ± 2.99c | 26.15 ± 1.50b | 0.04 ± 0.01a | 9.24 ± 0.11bc | 0.05 ± 0.00a | 0.99 ± 0.08b |
| C-DL | 7.71 ± 0.03a | 41.39 ± 1.37a | 20.89 ± 0.87a | 0.03 ± 0.01a | 4.72 ± 1.18ab | 0.05 ± 0.01a | 0.86 ± 0.04ab |
| C-HD | 7.96 ± 0.04b | 42.37 ± 0.18ab | 21.26 ± 0.18a | 0.04 ± 0.01a | 4.88 ± 0.31ab | 0.04 ± 0.00a | 1.06 ± 0.01b |
| C-SD | 7.92 ± 0.01ab | 48.87 ± 0.79bc | 24.13 ± 0.52ab | 0.03 ± 0.02a | 4.33 ± 0.93a | 0.05 ± 0.00a | 0.96 ± 0.08b |
| F | 8.96 | 6.53 | 5.98 | 0.46 | 5.99 | 0.55 | 5.69 |
| p | 0.017 | 0.032 | 0.038 | >0.05 | 0.038 | 0.709 | 0.42 |
| Site | Reads | OTU | Ace | Coverage | Chao | Shannon | Simpson |
|---|---|---|---|---|---|---|---|
| G-GH | 19,564 | 1365 | 1834 | 0.98 | 1823 | 5.67 | 0.012 |
| G-GF | 35,322 | 1419 | 1707 | 0.99 | 1739 | 5.64 | 0.011 |
| G-DL | 22,929 | 2219 | 2722 | 0.97 | 2767 | 6.58 | 0.004 |
| G-HD | 23,445 | 1695 | 2109 | 0.98 | 2066 | 5.74 | 0.017 |
| G-SD | 37,877 | 1500 | 1722 | 0.99 | 1715 | 4.15 | 0.131 |
| H-GH | 17,251 | 962 | 1347 | 0.98 | 1304 | 5.08 | 0.021 |
| H-GF | 24,239 | 1491 | 1885 | 0.98 | 1914 | 5.98 | 0.006 |
| H-DL | 30,447 | 1616 | 1957 | 0.99 | 1924 | 5.41 | 0.023 |
| H-HD | 21,065 | 1507 | 1928 | 0.98 | 1976 | 5.84 | 0.010 |
| H-SD | 29,558 | 1658 | 2171 | 0.98 | 2188 | 5.73 | 0.013 |
| C-GH | 19,755 | 1439 | 1906 | 0.98 | 1975 | 5.87 | 0.008 |
| C-GF | 31,367 | 1845 | 2220 | 0.99 | 2243 | 5.95 | 0.010 |
| C-DL | 25,934 | 1643 | 2172 | 0.98 | 2144 | 5.86 | 0.008 |
| C-HD | 19,231 | 1331 | 1731 | 0.98 | 1732 | 5.68 | 0.012 |
| C-SD | 26,614 | 1613 | 2113 | 0.98 | 2079 | 5.80 | 0.009 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, W.; Bahadur, A.; Sajjad, W.; Zhang, G.; Nasir, F.; Zhang, B.; Wu, X.; Liu, G.; Chen, T. Bacterial Diversity and Community Composition Distribution in Cold-Desert Habitats of Qinghai–Tibet Plateau, China. Microorganisms 2021, 9, 262. https://doi.org/10.3390/microorganisms9020262
Zhang W, Bahadur A, Sajjad W, Zhang G, Nasir F, Zhang B, Wu X, Liu G, Chen T. Bacterial Diversity and Community Composition Distribution in Cold-Desert Habitats of Qinghai–Tibet Plateau, China. Microorganisms. 2021; 9(2):262. https://doi.org/10.3390/microorganisms9020262
Chicago/Turabian StyleZhang, Wei, Ali Bahadur, Wasim Sajjad, Gaosen Zhang, Fahad Nasir, Binglin Zhang, Xiukun Wu, Guangxiu Liu, and Tuo Chen. 2021. "Bacterial Diversity and Community Composition Distribution in Cold-Desert Habitats of Qinghai–Tibet Plateau, China" Microorganisms 9, no. 2: 262. https://doi.org/10.3390/microorganisms9020262






