Longitudinal Microbiome Analysis in a Dextran Sulfate Sodium-Induced Colitis Mouse Model
Abstract
1. Introduction
2. Materials and Methods
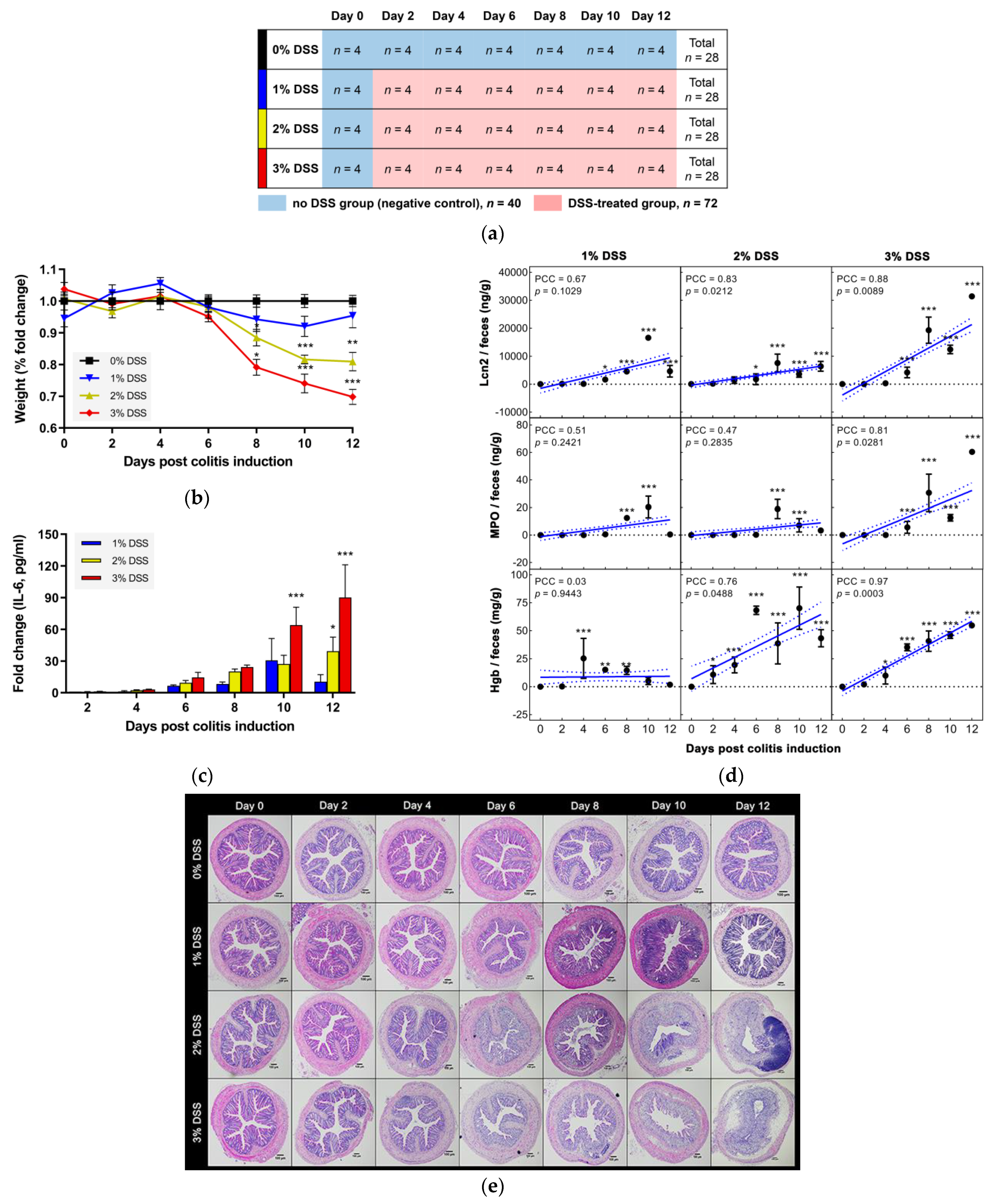
2.1. A Two-Factor Design for an Experimental Mouse in Vivo Colitis Model
2.2. Fecal Inflammatory Markers
2.3. Serum Inflammatory Cytokines
2.4. Histological Observations
2.5. DNA Extraction and Purification
2.6. 16S rRNA Gene Amplification and HiSeq Sequencing
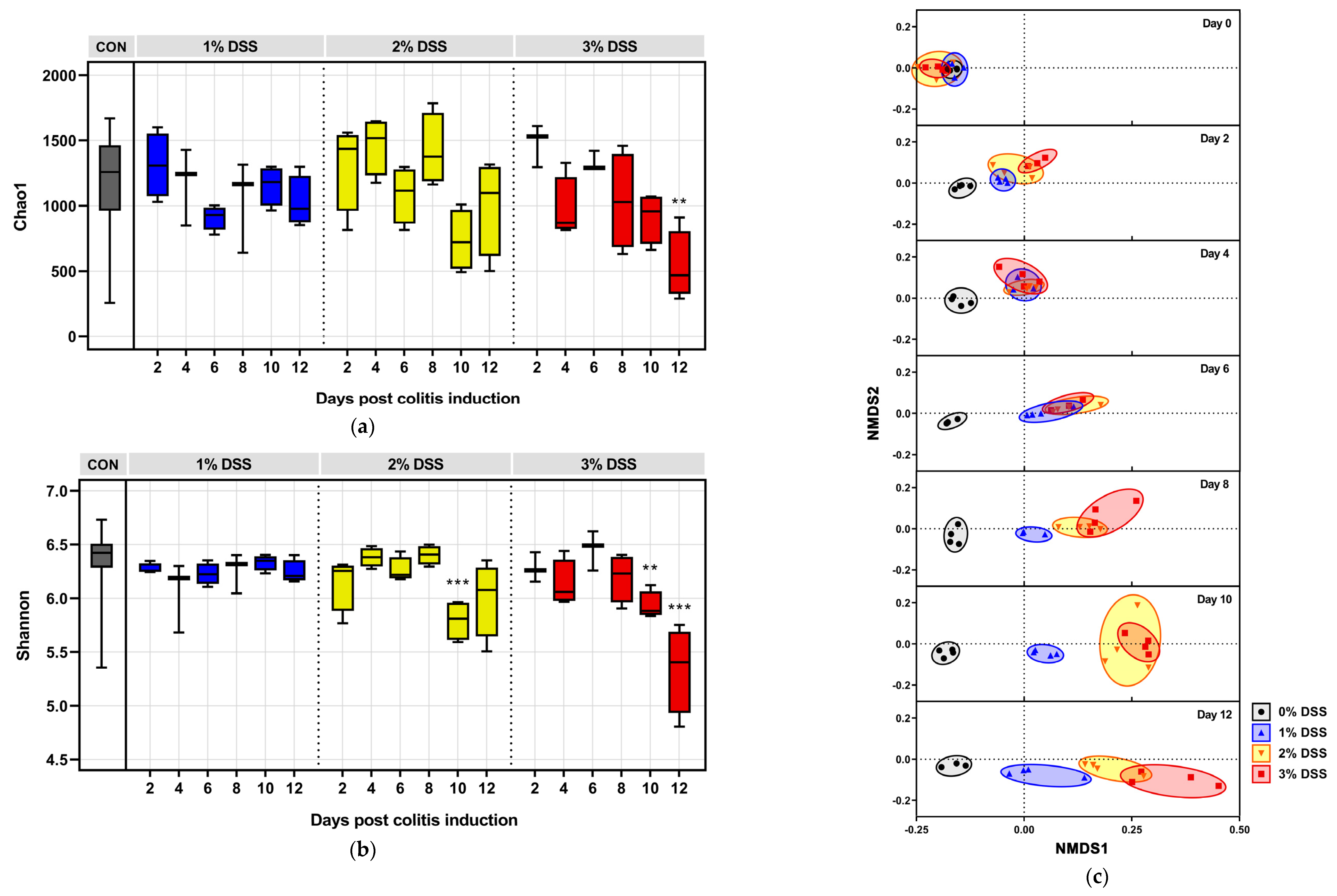
2.7. Data Processing, Taxonomic Classification, and Diversity Studies
2.8. Colitis-Associated Microbiota Analysis
2.9. Statistical Analyses
2.10. Data Availability
3. Results
3.1. DSS-Driven Colitis Development in a Two-Factor Designed in Vivo Mouse Model
3.2. DSS-Driven Microbial Diversity Shifts
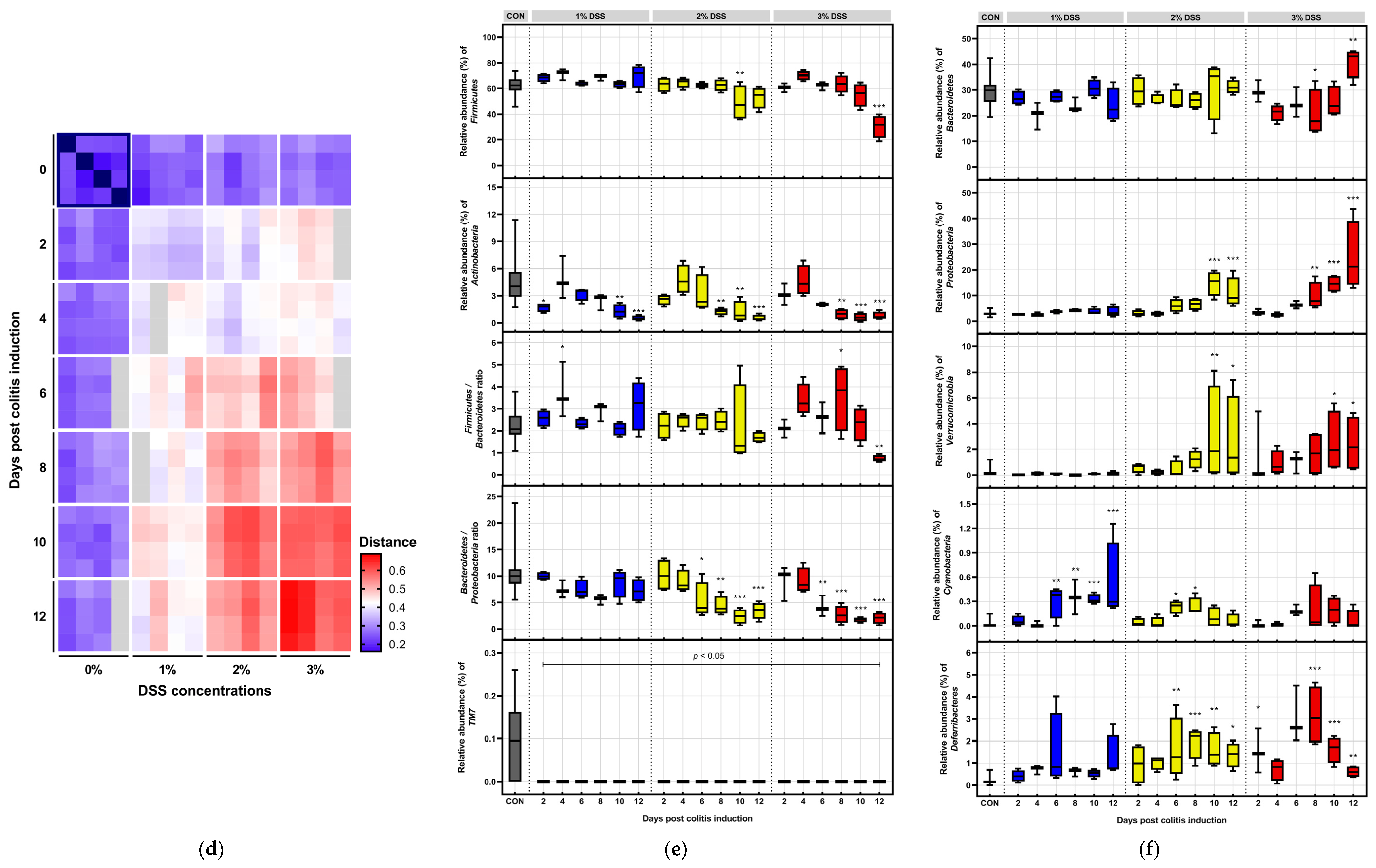
3.3. Microbial Dynamics Depending on DSS Exposure
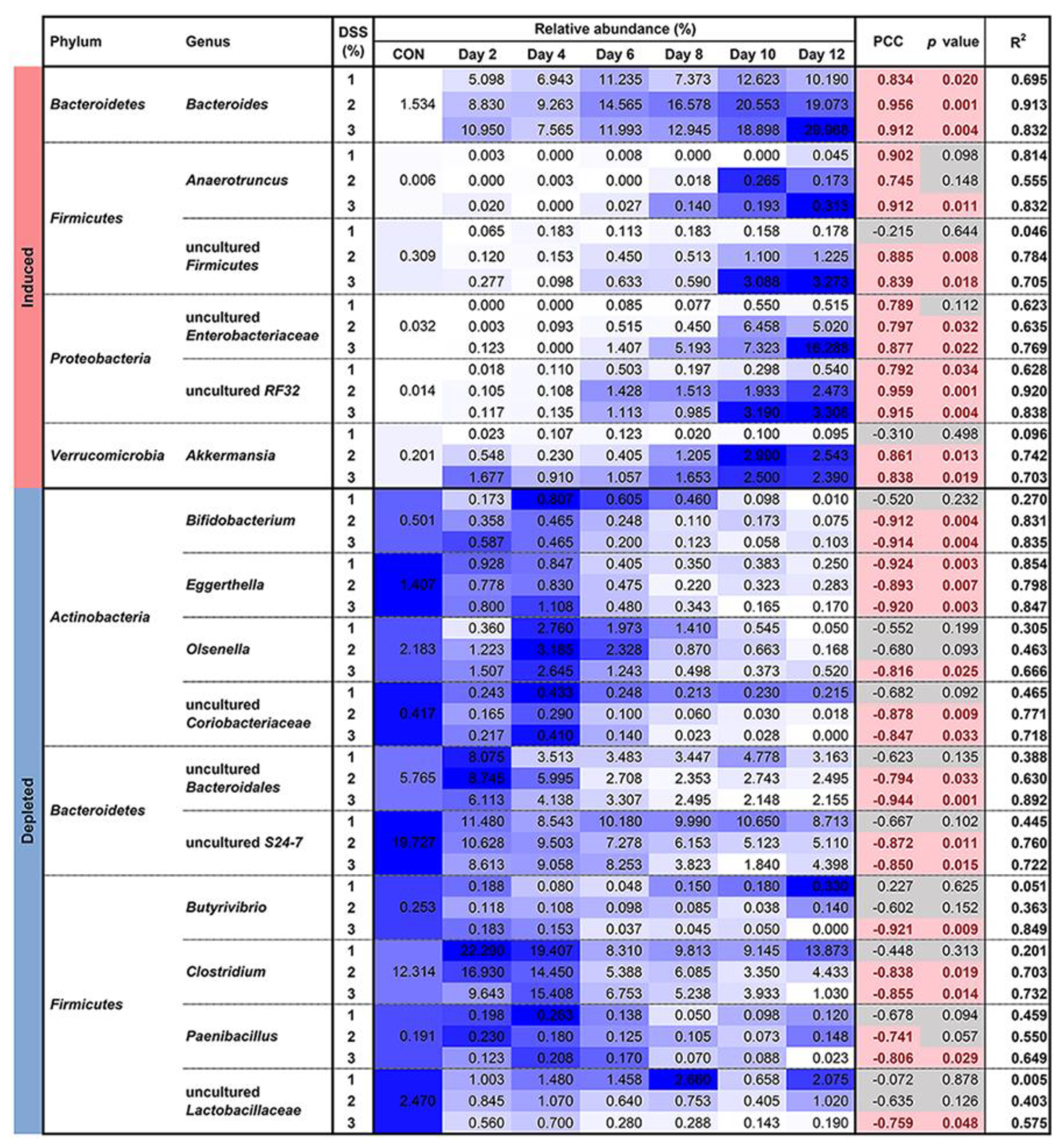
3.4. Microbial Susceptibility to Colitis Severity
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hold, G.L.; Smith, M.; Grange, C.; Watt, E.R.; El-Omar, E.M.; Mukhopadhya, I. Role of the gut microbiota in inflammatory bowel disease pathogenesis: What have we learnt in the past 10 years? World J. Gastroenterol. 2014, 20, 1192–1210. [Google Scholar] [CrossRef]
- Glassner, K.L.; Abraham, B.P.; Quigley, E.M.M. The microbiome and inflammatory bowel disease. J. Allergy Clin. Immunol. 2020, 145, 16–27. [Google Scholar] [CrossRef]
- Park, H.; Yeo, S.; Arellano, K.; Kim, H.R.; Holzapfel, W.H. Probiotics and Prebiotics in Animal Health and Food Safety; Di Gioia, D., Biavati, B., Eds.; Springer: Berlin, Germany, 2018; Chapter 2; pp. 35–62. ISBN 978-3-319-71950-4. [Google Scholar]
- Ni, J.; Wu, G.; Albenberg, L.; Tomov, V. Gut microbiota and IBD: Causation or correlation? Nat. Rev. Gastroenterol. Hepatol. 2017, 14, 573–584. [Google Scholar] [CrossRef]
- Caruso, R.; Lo, B.C.; Núñez, G. Host–microbiota interactions in inflammatory bowel disease. Nat. Rev. Immunol. 2020, 20, 411–426. [Google Scholar] [CrossRef]
- Roy, U.; Gálvez, E.J.C.; Iljazovic, A.; Lesker, T.R.; Błażejewski, A.J.; Pils, M.C.; Heise, U.; Huber, S.; Flavell, R.A.; Strowig, T. Distinct microbial communities trigger colitis development upon intestinal barrier damage via innate or adaptive immune cells. Cell Rep. 2017, 21, 994–1008. [Google Scholar] [CrossRef]
- Kitajima, S.; Morimoto, M.; Sagara, E.; Shimizu, C.; Ikeda, Y. Dextran sodium sulfate-induced colitis in germ-free IQI/Jic mice. Exp. Anim. 2001, 50, 387–395. [Google Scholar] [CrossRef]
- Du, Z.; Hudcovic, T.; Mrazek, J.; Kozakova, H.; Srutkova, D.; Schwarzer, M.; Tlaskalova-Hogenova, H.; Kostovcik, M.; Kverka, M. Development of gut inflammation in mice colonized with mucosa-associated bacteria from patients with ulcerative colitis. Gut Pathog. 2015, 7, 32. [Google Scholar] [CrossRef] [PubMed]
- Schirmer, M.; Garner, A.; Vlamakis, H.; Xavier, R.J. Microbial genes and pathways in inflammatory bowel disease. Nat. Rev. Microbiol. 2019, 17, 497–511. [Google Scholar] [CrossRef] [PubMed]
- Schlomann, B.H.; Parthasarathy, R. Timescales of gut microbiome dynamics. Curr. Opin. Microbiol. 2019, 50, 56–63. [Google Scholar] [CrossRef]
- Priya, S.; Blekhman, R. Population dynamics of the human gut microbiome: Change is the only constant. Genome Biol. 2019, 20, 150. [Google Scholar] [CrossRef] [PubMed]
- Halfvarson, J.; Brislawn, C.; Lamendella, R.; Vázquez-Baeza, Y.; Walters, W.A.; Bramer, L.M.; D’Amato, M.; Bonfiglio, F.; McDonald, D.; Gonzalez, A.; et al. Dynamics of the human gut microbiome in inflammatory bowel disease. Nat. Microbiol. 2017, 2, 17004. [Google Scholar] [CrossRef] [PubMed]
- Zuo, T.; Ng, S.C. The gut microbiota in the pathogenesis and therapeutics of inflammatory bowel disease. Front. Microbiol. 2018, 9, 2247. [Google Scholar] [CrossRef]
- Seksik, P. Microbiote intestinal et MICI [Gut microbiota and IBD]. Gastroenterol. Clin. Biol. 2010, 34, S48–S55. [Google Scholar] [CrossRef]
- Nishida, A.; Inoue, R.; Inatomi, O.; Bamba, S.; Naito, Y.; Andoh, A. Gut microbiota in the pathogenesis of inflammatory bowel disease. Clin. J. Gastroenterol. 2018, 11, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Alam, M.T.; Amos, G.C.A.; Murphy, A.R.J.; Murch, S.; Wellington, E.M.H.; Arasaradnam, R.P. Microbial imbalance in inflammatory bowel disease patients at different taxonomic levels. Gut Pathog. 2020, 12, 1. [Google Scholar] [CrossRef] [PubMed]
- Pittayanon, R.; Lau, J.T.; Leontiadis, G.I.; Tse, F.; Yuan, Y.; Surette, M.; Moayyedi, P. Differences in gut microbiota in patients with vs without inflammatory bowel diseases: A systematic review. Gastroenterology 2020, 158, 930–946.e1. [Google Scholar] [CrossRef]
- Imhann, F.; Vich Vila, A.; Bonder, M.J.; Fu, J.; Gevers, D.; Visschedijk, M.C.; Spekhorst, L.M.; Alberts, R.; Franke, L.; van Dullemen, H.M.; et al. Interplay of host genetics and gut microbiota underlying the onset and clinical presentation of inflammatory bowel disease. Gut 2018, 67, 108–119. [Google Scholar] [CrossRef]
- Galazzo, G.; Tedjo, D.I.; Wintjens, D.S.J.; Savelkoul, P.H.M.; Masclee, A.A.M.; Bodelier, A.G.L.; Pierik, M.J.; Jonkers, D.M.A.E.; Penders, J. Faecal microbiota dynamics and their relation to disease course in crohn’s disease. J. Crohns Colitis 2019, 13, 1273–1282. [Google Scholar] [CrossRef]
- DeVoss, J.; Diehl, L. Murine models of inflammatory bowel disease (IBD): Challenges of modeling human disease. Toxicol. Pathol. 2014, 42, 99–110. [Google Scholar] [CrossRef]
- Oh, S.Y.; Cho, K.A.; Kang, J.L.; Kim, K.H.; Woo, S.Y. Comparison of experimental mouse models of inflammatory bowel disease. Int. J. Mol. Med. 2014, 33, 333–340. [Google Scholar] [CrossRef]
- Kiesler, P.; Fuss, I.J.; Strober, W. Experimental models of inflammatory bowel diseases. Cell. Mol. Gastroenterol. Hepatol. 2015, 1, 154–170. [Google Scholar] [CrossRef] [PubMed]
- Laroui, H.; Ingersoll, S.A.; Liu, H.C.; Baker, M.T.; Ayyadurai, S.; Charania, M.A.; Laroui, F.; Yan, Y.; Sitaraman, S.V.; Merlin, D. Dextran sodium sulfate (DSS) induces colitis in mice by forming nano-lipocomplexes with medium-chain-length fatty acids in the colon. PLoS ONE 2012, 7, e32084. [Google Scholar] [CrossRef] [PubMed]
- Hans, W.; Schölmerich, J.; Gross, V.; Falk, W. The role of the resident intestinal flora in acute and chronic dextran sulfate sodium-induced colitis in mice. Eur. J. Gastroenterol. Hepatol. 2000, 12, 267–273. [Google Scholar] [CrossRef]
- Brinkman, B.M.; Becker, A.; Ayiseh, R.B.; Hildebrand, F.; Raes, J.; Huys, G.; Vandenabeele, P. Gut microbiota affects sensitivity to acute DSS-induced colitis independently of host genotype. Inflamm. Bowel Dis. 2013, 19, 2560–2567. [Google Scholar] [CrossRef]
- Nell, S.; Suerbaum, S.; Josenhans, C. The impact of the microbiota on the pathogenesis of IBD: Lessons from mouse infection models. Nat. Rev. Microbiol. 2010, 8, 564–577. [Google Scholar] [CrossRef]
- Eichele, D.D.; Kharbanda, K.K. Dextran sodium sulfate colitis murine model: An indispensable tool for advancing our understanding of inflammatory bowel diseases pathogenesis. World J. Gastroenterol. 2017, 23, 6016–6029. [Google Scholar] [CrossRef]
- Yeo, S.; Park, H.; Seo, E.; Kim, J.; Kim, B.K.; Choi, I.S.; Huh, C.S. Anti-inflammatory and gut microbiota modulatory effect of Lactobacillus rhamnosus strain LDTM 7511 in a dextran sulfate sodium-induced colitis murine model. Microorganisms 2020, 8, 845. [Google Scholar] [CrossRef] [PubMed]
- Bian, X.; Wu, W.; Yang, L.; Lv, L.; Wang, Q.; Li, Y.; Ye, J.; Fang, D.; Wu, J.; Jiang, X. Administration of Akkermansia muciniphila ameliorates dextran sulfate sodium-induced ulcerative colitis in mice. Front. Microbiol. 2019, 10, 2259. [Google Scholar] [CrossRef] [PubMed]
- Constante, M.; Fragoso, G.; Calvé, A.; Samba-Mondonga, M.; Santos, M.M. Dietary heme induces gut dysbiosis, aggravates colitis, and potentiates the development of adenomas in mice. Front. Microbiol. 2017, 8, 1809. [Google Scholar] [CrossRef]
- Nagalingam, N.A.; Kao, J.Y.; Young, V.B. Microbial ecology of the murine gut associated with the development of dextran sodium sulfate-induced colitis. Inflamm. Bowel Dis. 2011, 17, 917–926. [Google Scholar] [CrossRef]
- Huang, Y.L.; Chassard, C.; Hausmann, M.; von Itzstein, M.; Hennet, T. Sialic acid catabolism drives intestinal inflammation and microbial dysbiosis in mice. Nat. Commun. 2015, 6, 8141. [Google Scholar] [CrossRef]
- Berry, D.; Schwab, C.; Milinovich, G.; Reichert, J.; Ben Mahfoudh, K.; Decker, T.; Engel, M.; Hai, B.; Hainzl, E.; Heider, S.; et al. Phylotype-level 16S rRNA analysis reveals new bacterial indicators of health state in acute murine colitis. ISME J. 2012, 6, 2091–2106. [Google Scholar] [CrossRef]
- de Bruyn, M.; Sabino, J.; Vandeputte, D.; Vermeire, S.; Raes, J. Comparisons of gut microbiota profiles in wild-type and gelatinase B/matrix metalloproteinase-9-deficient mice in acute DSS-induced colitis. NPJ Biofilms Microbi. 2018, 4, 18. [Google Scholar] [CrossRef]
- Laukens, D.; Brinkman, B.M.; Raes, J.; De Vos, M.; Vandenabeele, P. Heterogeneity of the gut microbiome in mice: Guidelines for optimizing experimental design. FEMS Microbiol. Rev. 2016, 40, 117–132. [Google Scholar] [CrossRef] [PubMed]
- Hildebrand, F.; Nguyen, T.L.; Brinkman, B.; Yunta, R.G.; Cauwe, B.; Vandenabeele, P.; Liston, A.; Raes, J. Inflammation-associated enterotypes, host genotype, cage and inter-individual effects drive gut microbiota variation in common laboratory mice. Genome Biol. 2013, 14, R4. [Google Scholar] [CrossRef] [PubMed]
- Viennois, E.; Chen, F.; Laroui, H.; Baker, M.T.; Merlin, D. Dextran sodium sulfate inhibits the activities of both polymerase and reverse transcriptase: Lithium chloride purification, a rapid and efficient technique to purify RNA. BMC Res. Notes. 2013, 6, 360. [Google Scholar] [CrossRef] [PubMed]
- Kerr, T.A.; Ciorba, M.A.; Matsumoto, H.; Davis, V.R.; Luo, J.; Kennedy, S.; Xie, Y.; Shaker, A.; Dieckgraefe, B.K.; Davidson, N.O. Dextran sodium sulfate inhibition of real-time polymerase chain reaction amplification: A poly-A purification solution. Inflamm. Bowel Dis. 2012, 18, 344–348. [Google Scholar] [CrossRef]
- Green, M.R.; Sambrook, J. Precipitation of DNA with ethanol. Cold Spring Harb. Protoc. 2016. [Google Scholar] [CrossRef] [PubMed]
- Amplicon PCR, Clean-Up PCR, Index PCR. 16S Metagenomic Sequencing Library Preparation. Available online: https://www.illumina.com/content/dam/illumina-support/documents/documentation/chemistry_documentation/16s/16s-metagenomic-library-prep-guide-15044223-b.pdf (accessed on 18 October 2020).
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- McMurdie, P.J.; Holmes, S. phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef]
- Callahan, B.J.; Sankaran, K.; Fukuyama, J.A.; McMurdie, P.J.; Holmes, S.P. Bioconductor workflow for microbiome data analysis: From raw reads to community analyses. F1000Research 2016, 5, 1492. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Bittinger, K.; Charlson, E.S.; Hoffmann, C.; Lewis, J.; Wu, G.D.; Collman, R.G.; Bushman, F.D.; Li, H. Associating microbiome composition with environmental covariates using generalized UniFrac distances. Bioinformatics 2012, 28, 2106–2113. [Google Scholar] [CrossRef]
- Lagkouvardos, I.; Fischer, S.; Kumar, N.; Clavel, T. Rhea: A transparent and modular R pipeline for microbial profiling based on 16S rRNA gene amplicons. PeerJ 2017, 5, e2836. [Google Scholar] [CrossRef]
- Prata, M.M.; Havt, A.; Bolick, D.T.; Pinkerton, R.; Lima, A.; Guerrant, R.L. Comparisons between myeloperoxidase, lactoferrin, calprotectin and lipocalin-2, as fecal biomarkers of intestinal inflammation in malnourished children. J. Transl. Sci. 2016, 2, 134–139. [Google Scholar] [CrossRef] [PubMed]
- Chassaing, B.; Aitken, J.D.; Malleshappa, M.; Vijay-Kumar, M. Dextran sulfate sodium (DSS)-induced colitis in mice. Curr. Protoc. Immunol. 2014, 104, 15.25.1–15.25.14. [Google Scholar] [CrossRef]
- Solomon, L.; Mansor, S.; Mallon, P.; Donnelly, E.T.; Hoper, M.; Loughrey, M.; Kirk, S.; Gardiner, K. The dextran sulphate sodium (DSS) model of colitis: An overview. Comp. Clin. Pathol. 2010, 19, 235–239. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Anders, S.; Kim, V.; Huber, W. RNA-Seq workflow: Gene-level exploratory analysis and differential expression. F1000Research 2015, 4, 1070. [Google Scholar] [CrossRef] [PubMed]
- Soneson, C.; Delorenzi, M. A comparison of methods for differential expression analysis of RNA-seq data. BMC Bioinform. 2013, 14, 91. [Google Scholar] [CrossRef] [PubMed]
- Khan, I.; Ullah, N.; Zha, L.; Bai, Y.; Khan, A.; Zhao, T.; Che, T.; Zhang, C. Alteration of gut microbiota in inflammatory bowel disease (IBD): Cause or consequence? IBD treatment targeting the gut microbiome. Pathogens 2019, 8, 126. [Google Scholar] [CrossRef]
- Toor, D.; Wsson, M.K.; Kumar, P.; Karthikeyan, G.; Kaushik, N.K.; Goel, C.; Singh, S.; Kumar, A.; Prakash, H. Dysbiosis disrupts gut immune homeostasis and promotes gastric diseases. Int. J. Mol. Sci. 2019, 20, 2432. [Google Scholar] [CrossRef] [PubMed]
- Perše, M.; Cerar, A. Dextran sodium sulphate colitis mouse model: Traps and tricks. J. Biomed. Biotechnol. 2012, 2012, 718617. [Google Scholar] [CrossRef]
- Li, M.; Wu, Y.; Hu, Y.; Zhao, L.; Zhang, C. Initial gut microbiota structure affects sensitivity to DSS-induced colitis in a mouse model. Sci. China Life Sci. 2018, 61, 762–769. [Google Scholar] [CrossRef]
- Sharpton, T.; Lyalina, S.; Luong, J.; Pham, J.; Deal, E.M.; Armour, C.; Gaulke, C.; Sanjabi, S.; Pollard, K.S. Development of inflammatory bowel disease is linked to a longitudinal restructuring of the gut metagenome in mice. mSystems 2017, 2, e00036-17. [Google Scholar] [CrossRef] [PubMed]
- Schwab, C.; Berry, D.; Rauch, I.; Rennisch, I.; Ramesmayer, J.; Hainzl, E.; Heider, S.; Decker, T.; Kenner, L.; Müller, M.; et al. Longitudinal study of murine microbiota activity and interactions with the host during acute inflammation and recovery. ISME J. 2014, 8, 1101–1114. [Google Scholar] [CrossRef]
- Zeng, M.Y.; Inohara, N.; Nuñez, G. Mechanisms of inflammation-driven bacterial dysbiosis in the gut. Mucosal Immunol. 2017, 10, 18–26. [Google Scholar] [CrossRef]
- Robinson, L.S.; Warren, G.L.; Amanda, L.L. The sialate O-acetylesterase EstA from gut Bacteroidetes species enables sialidase-mediated cross-species foraging of 9-O-acetylated sialoglycans. J. Biol. Chem. 2017, 292, 11861–11872. [Google Scholar] [CrossRef]
- Singh, V.; Yeoh, B.S.; Xiao, X.; Kumar, M.; Bachman, M.; Borregaard, N.; Joe, B.; Vijay-Kumar, M. Interplay between enterobactin, myeloperoxidase and lipocalin 2 regulates E. coli survival in the inflamed gut. Nat. Commun. 2015, 6, 7113. [Google Scholar] [CrossRef]
- Bachman, M.A.; Miller, V.L.; Weiser, J.N. Mucosal lipocalin 2 has pro-inflammatory and iron-sequestering effects in response to bacterial enterobactin. PLoS Pathog. 2009, 5, e1000622. [Google Scholar] [CrossRef] [PubMed]
- Ijssennagger, N.; Belzer, C.; Hooiveld, G.J.; Dekker, J.; van Mil, S.W.; Müller, M.; Kleerebezem, M.; van der Meer, R. Gut microbiota facilitates dietary heme-induced epithelial hyperproliferation by opening the mucus barrier in colon. Proc. Natl. Acad. Sci. USA 2015, 112, 10038–10043. [Google Scholar] [CrossRef]
- Thompson, C.; Mikaelyan, A.; Brune, A. Immune-modulating gut symbionts are not “Candidatus Arthromitus”. Mucosal Immunol. 2013, 6, 200–201. [Google Scholar] [CrossRef]
- Lécuyer, E.; Rakotobe, S.; Lengliné-Garnier, H.; Lebreton, C.; Picard, M.; Juste, C.; Fritzen, R.; Eberl, G.; McCoy, K.D.; Macpherson, A.J.; et al. Segmented filamentous bacterium uses secondary and tertiary lymphoid tissues to induce gut IgA and specific T helper 17 cell responses. Immunity 2014, 40, 608–620. [Google Scholar] [CrossRef]
- Goto, Y.; Panea, C.; Nakato, G.; Cebula, A.; Lee, C.; Diez, M.G.; Laufer, T.M.; Ignatowicz, L.; Ivanov, I.I. Segmented filamentous bacteria antigens presented by intestinal dendritic cells drive mucosal Th17 cell differentiation. Immunity 2014, 40, 594–607. [Google Scholar] [CrossRef]
- Gaboriau-Routhiau, V.; Rakotobe, S.; Lécuyer, E.; Mulder, I.; Lan, A.; Bridonneau, C.; Rochet, V.; Pisi, A.; De Paepe, M.; Brandi, G.; et al. The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity 2009, 31, 677–689. [Google Scholar] [CrossRef] [PubMed]
- Ash, C.; Priest, F.G.; Collins, M.D. Molecular identification of rRNA group 3 bacilli (Ash, Farrow, Wallbanks and Collins) using a PCR probe test. Proposal for the creation of a new genus Paenibacillus. Antonie Van Leeuwenhoek 1993, 64, 253–260. [Google Scholar] [CrossRef]
- Lorentz, R.H.; Artico, S.; da Silveira, A.B.; Einsfeld, A.; Corção, G. Evaluation of antimicrobial activity in Paenibacillus spp. strains isolated from natural environment. Lett. Appl. Microbiol. 2006, 43, 541–547. [Google Scholar] [CrossRef] [PubMed]
- Bao, G.H.; Ho, C.T.; Barasch, J. The ligands of neutrophil gelatinase-associated lipocalin. RSC Adv. 2015, 5, 104363–104374. [Google Scholar] [CrossRef]
- Loubinoux, J.; Bronowicki, J.P.; Pereira, I.A.; Mougenel, J.L.; Faou, A.E. Sulfate-reducing bacteria in human feces and their association with inflammatory bowel diseases. FEMS Microbiol. Ecol. 2002, 40, 107–112. [Google Scholar] [CrossRef]
- Kushkevych, I.; Dordević, D.; Kollar, P.; Vítězová, M.; Drago, L. Hydrogen sulfide as a toxic product in the small-large intestine axis and its role in IBD development. J. Clin. Med. 2019, 8, 1054. [Google Scholar] [CrossRef] [PubMed]
- Coutinho, C.M.L.M.; Coutinho-Silva, R.; Zinkevich, V.; Pearce, C.B.; Ojcius, D.M.; Beech, I. Sulphate-reducing bacteria from ulcerative colitis patients induce apoptosis of gastrointestinal epithelial cells. Microb. Pathog. 2017, 112, 126–134. [Google Scholar] [CrossRef]
- Dordević, D.; Jančíková, S.; Vítězová, M.; Kushkevych, I. Hydrogen sulfide toxicity in the gut environment: Meta-analysis of sulfate-reducing and lactic acid bacteria in inflammatory processes. J. Adv. Res. 2020, 27, 55–69. [Google Scholar] [CrossRef] [PubMed]
- Håkansson, Å.; Tormo-Badia, N.; Baridi, A.; Xu, J.; Molin, G.; Hagslätt, M.L.; Karlsson, C.; Jeppsson, B.; Cilio, C.M.; Ahrné, S. Immunological alteration and changes of gut microbiota after dextran sulfate sodium (DSS) administration in mice. Clin. Exp. Med. 2015, 15, 107–120. [Google Scholar] [CrossRef]
- Yang, Y.; Chen, G.; Yang, Q.; Ye, J.; Cai, X.; Tsering, P.; Cheng, X.; Hu, C.; Zhang, S.; Cao, P. Gut microbiota drives the attenuation of dextran sulphate sodium-induced colitis by Huangqin decoction. Oncotarget 2017, 8, 48863–48874. [Google Scholar] [CrossRef]
- Ding, S.; Ma, Y.; Liu, G.; Yan, W.; Jiang, H.; Fang, J. Lactobacillus brevis alleviates DSS-induced colitis by reprograming intestinal microbiota and influencing serum metabolome in murine model. Front. Physiol. 2019, 10, 1152. [Google Scholar] [CrossRef]
- Anyansi, C.; Straub, T.J.; Manson, A.L.; Earl, A.M.; Abeel, T. Computational methods for strain-level microbial detection in colony and metagenome sequencing data. Front. Microbiol. 2020, 11, 1925. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Nguyen, L.H.; Franzosa, E.A.; Huttenhower, C. Strain-level epidemiology of microbial communities and the human microbiome. Genome Med. 2020, 12, 71. [Google Scholar] [CrossRef]
- Thomas, A.M.; Segata, N. Multiple levels of the unknown in microbiome research. BMC Biol. 2019, 17, 48. [Google Scholar] [CrossRef] [PubMed]
- Vrancken, G.; Gregory, A.C.; Huys, G.R.B.; Faust, K.; Raes, J. Synthetic ecology of the human gut microbiota. Nat. Rev. Microbiol. 2019, 17, 754–763. [Google Scholar] [CrossRef]
- Lagier, J.C.; Hugon, P.; Khelaifia, S.; Fournier, P.E.; La Scola, B.; Raoult, D. The rebirth of culture in microbiology through the example of culturomics to study human gut microbiota. Clin. Microbiol. Rev. 2015, 28, 237–264. [Google Scholar] [CrossRef]
- Kambouris, M.E.; Pavlidis, C.; Skoufas, E.; Arabatzis, M.; Kantzanou, M.; Velegraki, A.; Patrinos, G.P. Culturomics: A new kid on the block of OMICS to enable personalized medicine. OMICS 2018, 22, 108–118. [Google Scholar] [CrossRef] [PubMed]
- Cross, K.L.; Campbell, J.H.; Balachandran, M.; Campbell, A.G.; Cooper, S.J.; Griffen, A.; Heaton, M.; Joshi, S.; Klingeman, D.; Leys, E.; et al. Targeted isolation and cultivation of uncultivated bacteria by reverse genomics. Nat. Biotechnol. 2019, 37, 1314–1321. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Park, H.; Yeo, S.; Kang, S.; Huh, C.S. Longitudinal Microbiome Analysis in a Dextran Sulfate Sodium-Induced Colitis Mouse Model. Microorganisms 2021, 9, 370. https://doi.org/10.3390/microorganisms9020370
Park H, Yeo S, Kang S, Huh CS. Longitudinal Microbiome Analysis in a Dextran Sulfate Sodium-Induced Colitis Mouse Model. Microorganisms. 2021; 9(2):370. https://doi.org/10.3390/microorganisms9020370
Chicago/Turabian StylePark, Hyunjoon, Soyoung Yeo, Seokwon Kang, and Chul Sung Huh. 2021. "Longitudinal Microbiome Analysis in a Dextran Sulfate Sodium-Induced Colitis Mouse Model" Microorganisms 9, no. 2: 370. https://doi.org/10.3390/microorganisms9020370
APA StylePark, H., Yeo, S., Kang, S., & Huh, C. S. (2021). Longitudinal Microbiome Analysis in a Dextran Sulfate Sodium-Induced Colitis Mouse Model. Microorganisms, 9(2), 370. https://doi.org/10.3390/microorganisms9020370





