Regulatory Control of Rishirilide(s) Biosynthesis in Streptomyces bottropensis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains, Plasmids, and Growth Conditions
2.2. Generation of Gene Deletion Mutants
2.3. Construction of the Plasmids for Complementation and Overexpression
2.4. Strain Fermentation and Analysis of Rishirilides Production
2.5. Total RNA Isolation and RT-PCR
2.6. Gus-Assay
2.7. Production and Purification of RslR3DBD
2.8. Production and Purification of RslR4
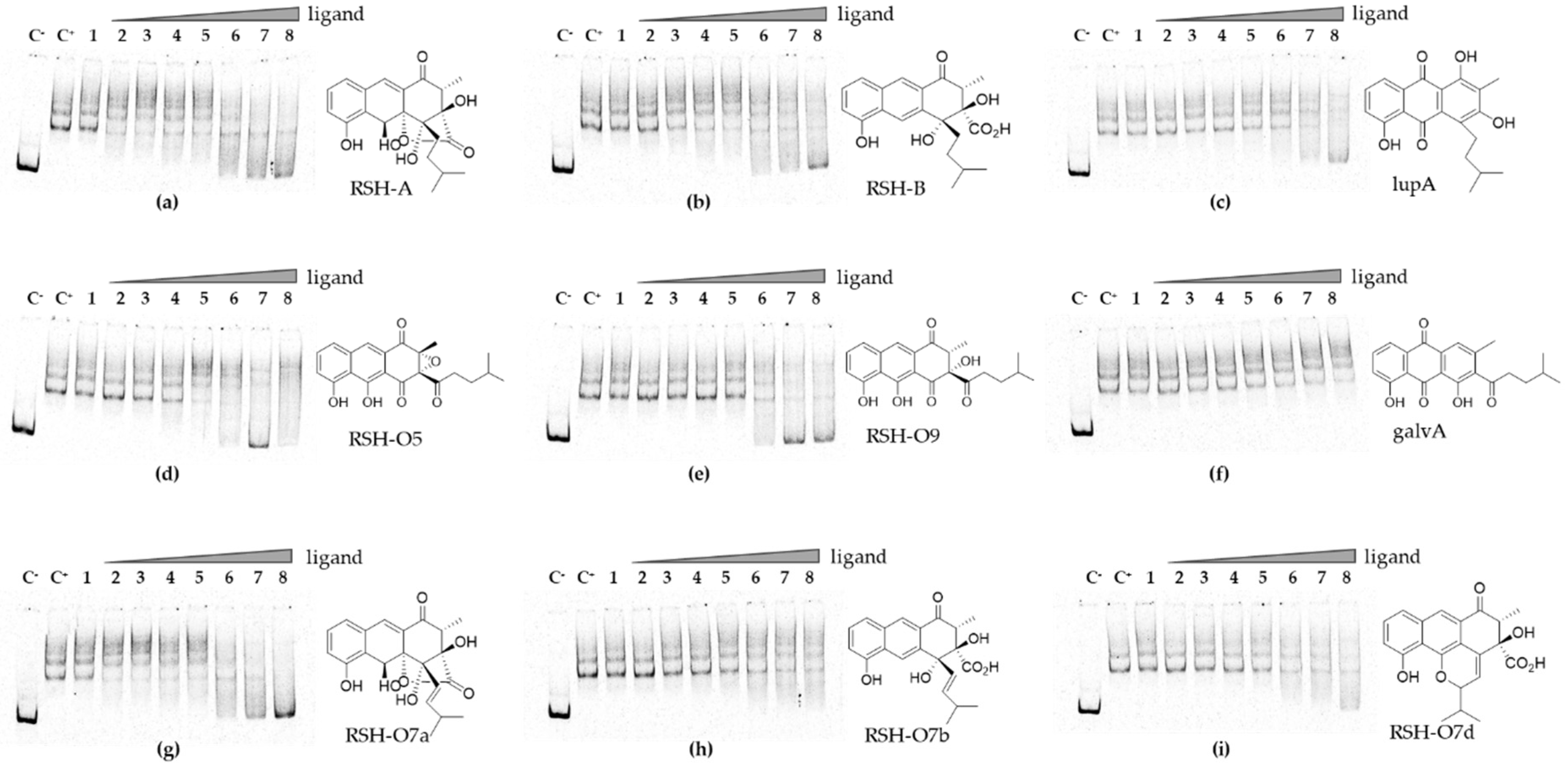
2.9. Electrophoretic Mobility Shift Assay
3. Results
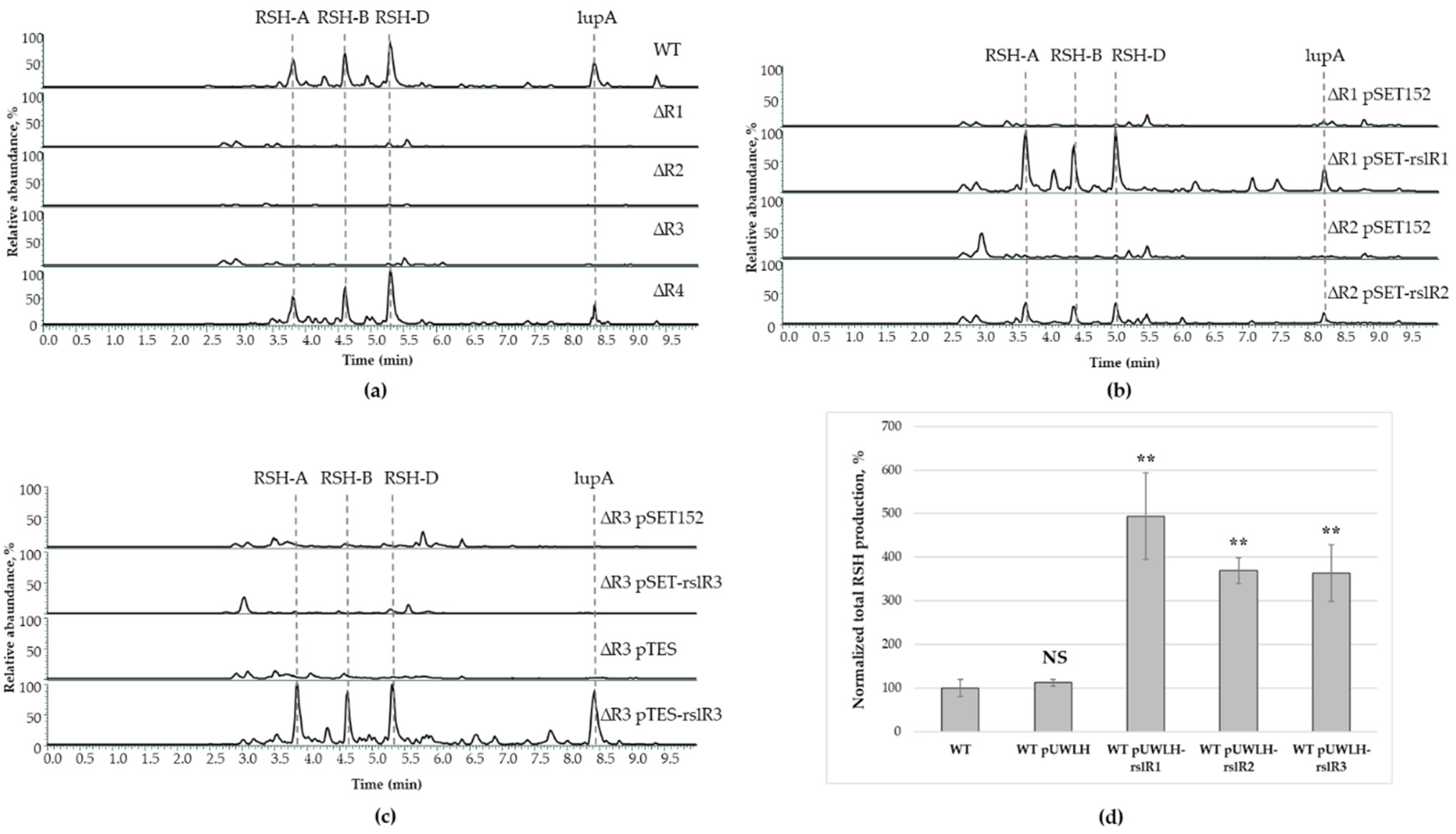
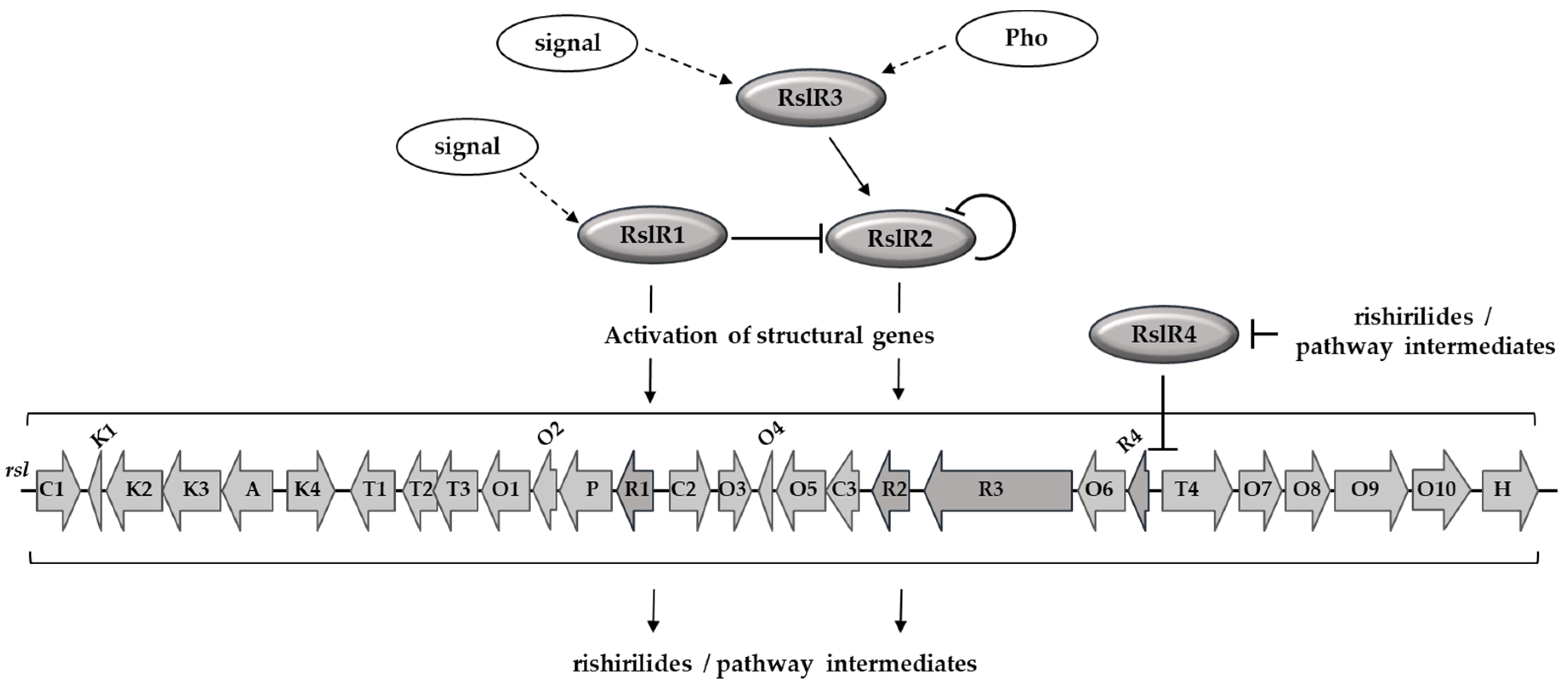
3.1. Biosynthesis of Rishirilides Is Positively Regulated by RslR1, RslR2, and RslR3 Cluster-Situated Regulators
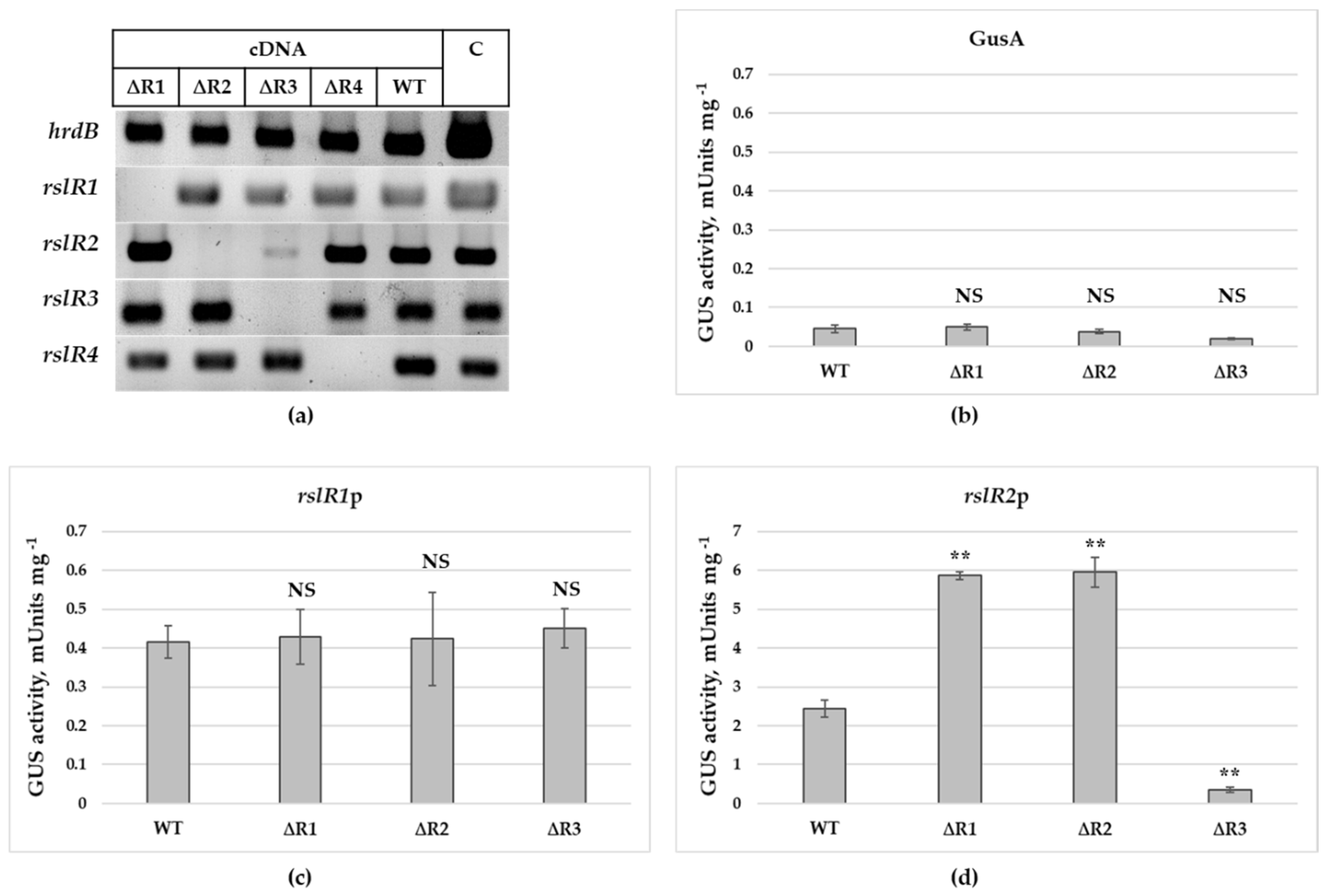
3.2. Expression Pattern of Regulatory Genes in Wild-Type and Mutant Strains
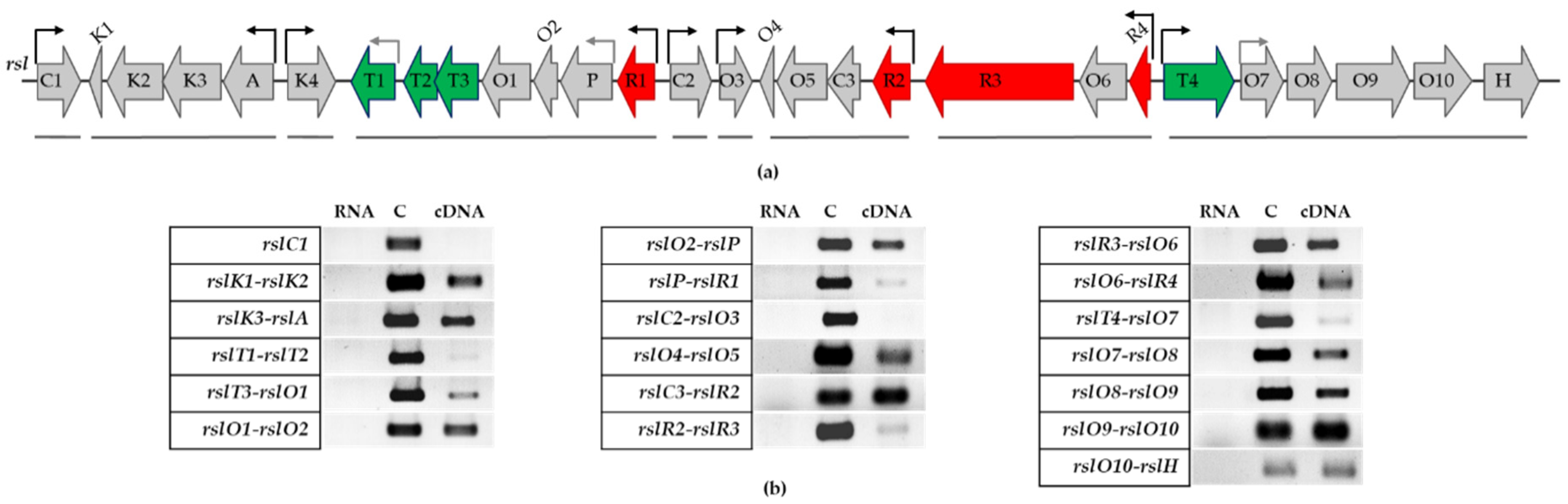
3.3. Transcriptional Organization of rsl-Genes
3.4. RslR3 is a Multi-Domain Protein That Regulates rslR2
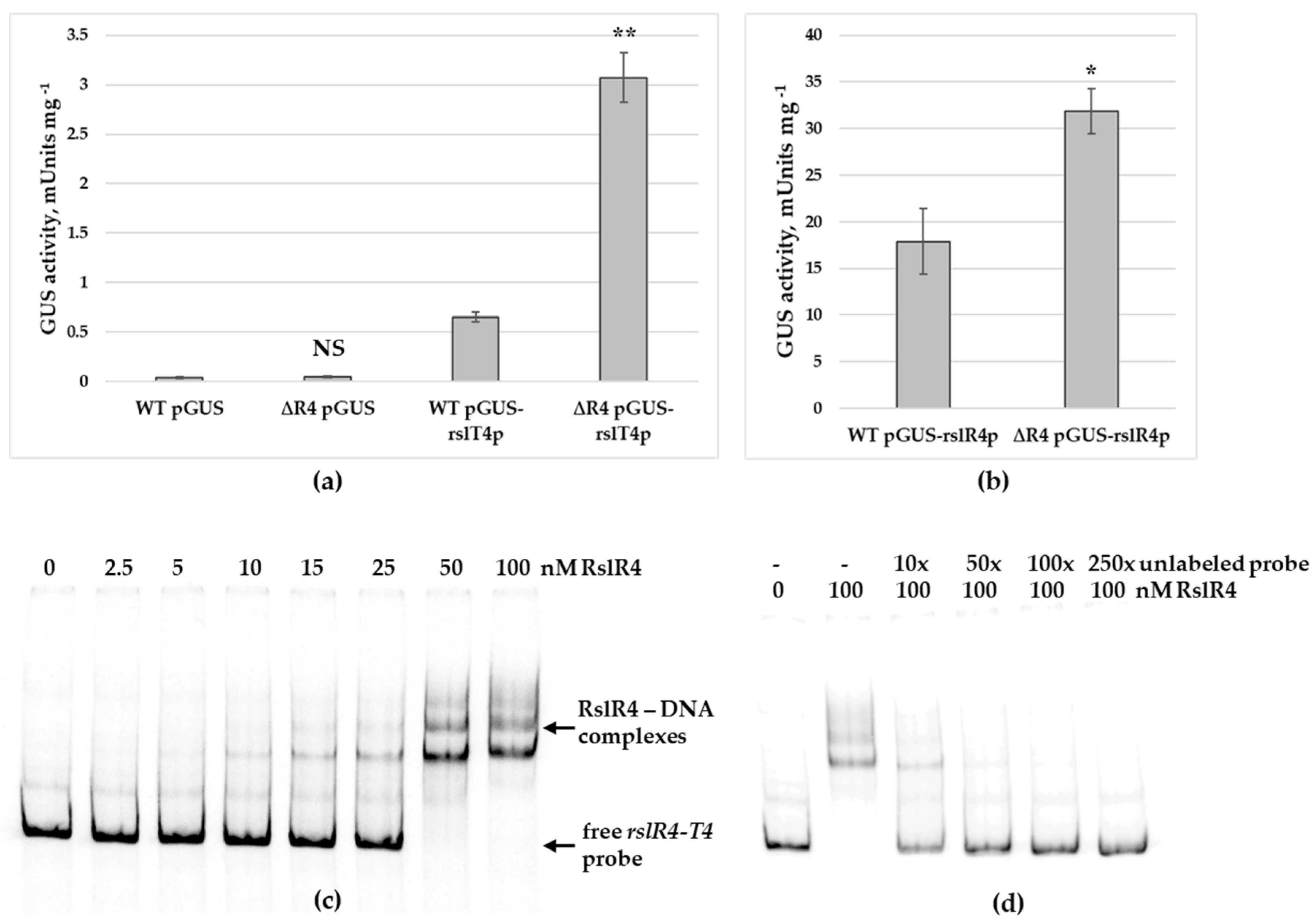
3.5. RslR4 Negatively Regulates Transcription of MFS Transporter Gene rslT4 and Its Own Expression
3.6. Rishirilides and Intermediates of the Pathway Interact with RslR4
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Flärdh, K.; Buttner, M.J. Streptomyces morphogenetics: Dissecting differentiation in a filamentous bacterium. Nat. Rev. Genet. 2009, 7, 36–49. [Google Scholar] [CrossRef]
- Quinn, G.A.; Banat, A.M.; Abdelhameed, A.M.; Banat, I.M. Streptomyces from traditional medicine: Sources of new innovations in antibiotic discovery. J. Med. Microbiol. 2020, 69, 1040–1048. [Google Scholar] [CrossRef]
- Procópio, R.E.D.L.; Da Silva, I.R.; Martins, M.K.; De Azevedo, J.L.; De Araújo, J.M. Antibiotics produced by Streptomyces. Braz. J. Infect. Dis. 2012, 16, 466–471. [Google Scholar] [CrossRef] [Green Version]
- Sivalingam, P.; Hong, K.; Pote, J.; Prabakar, K. Extreme Environment Streptomyces: Potential Sources for New Antibacterial and Anticancer Drug Leads? Int. J. Microbiol. 2019, 2019, 5283948. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsypik, O.; Makitrynskyy, R.; Bera, A.; Song, L.; Wohlleben, W.; Fedorenko, V.; Ostash, B. Role of GntR Family Regulatory Gene SCO1678 in Gluconate Metabolism in Streptomyces coelicolor M145. BioMed Res. Int. 2017, 2017, 9529501. [Google Scholar] [CrossRef]
- Liu, G.; Chater, K.F.; Chandra, G.; Niu, G.; Tan, H. Molecular Regulation of Antibiotic Biosynthesis in Streptomyces. Microbiol. Mol. Biol. Rev. 2013, 77, 112–143. [Google Scholar] [CrossRef] [Green Version]
- Xia, H.; Li, X.; Li, Z.; Zhan, X.; Mao, X.; Li, Y. The Application of Regulatory Cascades in Streptomyces: Yield Enhancement and Metabolite Mining. Front. Microbiol. 2020, 11, 406. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martín, J.F.; Liras, P. The Balance Metabolism Safety Net: Integration of Stress Signals by Interacting Transcriptional Factors in Streptomyces and Related Actinobacteria. Front. Microbiol. 2020, 10, 3120. [Google Scholar] [CrossRef]
- Kong, D.; Wang, X.; Nie, J.; Niu, G. Regulation of Antibiotic Production by Signaling Molecules in Streptomyces. Front. Microbiol. 2019, 10, 2927. [Google Scholar] [CrossRef] [Green Version]
- Hesketh, A.; Chen, W.; Ryding, J.; Chang, S.; Bibb, M. The global role of ppGpp synthesis in morphological differentiation and antibiotic production in Streptomyces coelicolor A3(2). Genome Biol. 2007, 8, R161. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gomez-Escribano, J.P.; Martín, J.F.; Hesketh, A.; Bibb, M.J.; Liras, P. Streptomyces clavuligerus relA-null mutants overproduce clavulanic acid and cephamycin C: Negative regulation of secondary metabolism by (p)ppGpp. Microbiology 2008, 154, 744–755. [Google Scholar] [CrossRef] [Green Version]
- Hesketh, A.; Sun, J.; Bibb, M.J. Induction of ppGpp synthesis in Streptomyces coelicolor A3(2) grown under conditions of nutritional sufficiency elicits actII-ORF4 transcription and actinorhodin biosynthesis. Mol. Microbiol. 2001, 39, 136–144. [Google Scholar] [CrossRef] [Green Version]
- Nuzzo, D.; Makitrynskyy, R.; Tsypik, O.; Bechthold, A. Cyclic di-GMP cyclase SSFG_02181 from Streptomyces ghanaensis ATCC14672 regulates antibiotic biosynthesis and morphological differentiation in streptomycetes. Sci. Rep. 2020, 10, 12021. [Google Scholar] [CrossRef]
- Makitrynskyy, R.; Tsypik, O.; Nuzzo, D.; Paululat, T.; Zechel, D.L.; Bechthold, A. Secondary nucleotide messenger c-di-GMP exerts a global control on natural product biosynthesis in streptomycetes. Nucleic Acids Res. 2020, 48, 1583–1598. [Google Scholar] [CrossRef] [Green Version]
- Nuzzo, D.; Makitrynskyy, R.; Tsypik, O.; Bechthold, A. Identification and Characterization of Four c-di-GMP-Metabolizing Enzymes from Streptomyces ghanaensis ATCC14672 Involved in the Regulation of Morphogenesis and Moenomycin A Biosynthesis. Microorganisms 2021, 9, 284. [Google Scholar] [CrossRef]
- Nett, M.; Ikeda, H.; Moore, B.S. Genomic basis for natural product biosynthetic diversity in the actinomycetes. Nat. Prod. Rep. 2009, 26, 1362–1384. [Google Scholar] [CrossRef] [PubMed]
- Bate, N.; Stratigopoulos, G.; Cundliffe, E. Differential roles of two SARP-encoding regulatory genes during tylosin biosynthesis. Mol. Microbiol. 2002, 43, 449–458. [Google Scholar] [CrossRef]
- Bignell, D.R.D.; Bate, N.; Cundliffe, E. Regulation of tylosin production: Role of a TylP-interactive ligand. Mol. Microbiol. 2007, 63, 838–847. [Google Scholar] [CrossRef]
- Li, R.; Liu, G.; Xie, Z.; He, X.; Chen, W.; Deng, Z.; Tan, H. PolY, a transcriptional regulator with ATPase activity, directly activates transcription ofpolRin polyoxin biosynthesis inStreptomyces cacaoi. Mol. Microbiol. 2010, 75, 349–364. [Google Scholar] [CrossRef] [PubMed]
- Ostash, B.; Saghatelian, A.; Walker, S. A Streamlined Metabolic Pathway for the Biosynthesis of Moenomycin A. Chem. Biol. 2007, 14, 257–267. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bibb, M.J. Regulation of secondary metabolism in streptomycetes. Curr. Opin. Microbiol. 2005, 8, 208–215. [Google Scholar] [CrossRef]
- Gramajo, H.C.; Takano, E.; Bibb, M.J. Stationary-phase production of the antibiotic actinorhodin in Streptomyces coelicolor A3(2) is transcriptionally regulated. Mol. Microbiol. 1993, 7, 837–845. [Google Scholar] [CrossRef] [Green Version]
- Arias, P.; Fernández-Moreno, M.A.; Malpartida, F. Characterization of the Pathway-Specific Positive Transcriptional Regulator for Actinorhodin Biosynthesis inStreptomyces coelicolor A3(2) as a DNA-Binding Protein. J. Bacteriol. 1999, 181, 6958–6968. [Google Scholar] [CrossRef] [Green Version]
- Fujii, T.; Gramajo, H.C.; Takano, E.; Bibb, M.J. redD and actII-ORF4, pathway-specific regulatory genes for antibiotic production in Streptomyces coelicolor A3(2), are transcribed in vitro by an RNA polymerase holoenzyme containing sigma hrdD. J. Bacteriol. 1996, 178, 3402–3405. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- White, J.; Bibb, M. bldA dependence of undecylprodigiosin production in Streptomyces coelicolor A3(2) involves a pathway-specific regulatory cascade. J. Bacteriol. 1997, 179, 627–633. [Google Scholar] [CrossRef] [Green Version]
- Wolański, M.; Donczew, R.; Kois-Ostrowska, A.; Masiewicz, P.; Jakimowicz, D.; Zakrzewska-Czerwińska, J. The Level of AdpA Directly Affects Expression of Developmental Genes in Streptomyces coelicolor. J. Bacteriol. 2011, 193, 6358–6365. [Google Scholar] [CrossRef] [Green Version]
- Rigali, S.; Titgemeyer, F.; Barends, S.; Mulder, S.; Thomae, A.W.; Hopwood, D.A.; Van Wezel, G.P. Feast or famine: The global regulator DasR links nutrient stress to antibiotic production by Streptomyces. EMBO Rep. 2008, 9, 670–675. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, R.; Mast, Y.; Wang, J.; Zhang, W.; Zhao, G.; Wohlleben, W.; Lu, Y.; Jiang, W.-H. Identification of two-component system AfsQ1/Q2 regulon and its cross-regulation with GlnR inStreptomyces coelicolor. Mol. Microbiol. 2012, 87, 30–48. [Google Scholar] [CrossRef] [PubMed]
- McKenzie, N.L.; Nodwell, J.R. Phosphorylated AbsA2 Negatively Regulates Antibiotic Production in Streptomyces coelicolor through Interactions with Pathway-Specific Regulatory Gene Promoters. J. Bacteriol. 2007, 189, 5284–5292. [Google Scholar] [CrossRef] [Green Version]
- Yan, X.; Probst, K.; Linnenbrink, A.; Arnold, M.; Paululat, T.; Zeeck, A.; Bechthold, A. Cloning and Heterologous Expression of Three Type II PKS Gene Clusters fromStreptomyces bottropensis. ChemBioChem 2011, 13, 224–230. [Google Scholar] [CrossRef] [PubMed]
- Iwaki, H.; Nakayama, Y.; Takahashi, M.; Uetsuki, S.; Kido, M.; Fukuyama, Y. Structures of rishirilides A and B, alpha 2-macroglobulin inhibitors produced by Streptomyces rishiriensis OFR-1056. J. Antibiot. 1984, 37, 1091–1093. [Google Scholar] [CrossRef] [Green Version]
- Komagata, D.; Sawa, R.; Kinoshita, N.; Imada, C.; Sawa, T.; Naganawa, H.; Hamada, M.; Okami, Y.; Takeuchi, T. Isolation of glutathione-S- transferase inhibitors. J. Antibiot. 1992, 45, 1681–1683. [Google Scholar] [CrossRef] [Green Version]
- Schwarzer, P.; Tsypik, O.; Zuo, C.; Alali, A.; Wunsch-Palasis, J.; Heitzler, T.; Derochefort, J.; Bernhardt, M.; Yan, X.; Paululat, T.; et al. Early Steps in the Biosynthetic Pathway of Rishirilide B. Molecules 2020, 25, 1955. [Google Scholar] [CrossRef] [Green Version]
- Schwarzer, P.; Wunsch-Palasis, J.; Bechthold, A.; Paululat, T. Biosynthesis of Rishirilide B. Antibiotics 2018, 7, 20. [Google Scholar] [CrossRef] [Green Version]
- Tsypik, O.; Makitrynskyy, R.; Frensch, B.; Zechel, D.L.; Paululat, T.; Teufel, R.; Bechthold, A. Oxidative Carbon Backbone Rearrangement in Rishirilide Biosynthesis. J. Am. Chem. Soc. 2020, 142, 5913–5917. [Google Scholar] [CrossRef]
- Ostash, B.; Rebets, Y.; Myronovskyy, M.; Tsypik, O.; Ostash, I.; Kulachkovskyy, O.; Datsyuk, Y.; Nakamura, T.; Walker, S.; Fedorenko, V. Identification and characterization of the Streptomyces globisporus 1912 regulatory gene lndYR that affects sporulation and antibiotic production. Microbiology 2011, 157, 1240–1249. [Google Scholar] [CrossRef] [Green Version]
- Gust, B.; Chandra, G.; Jakimowicz, D.; Yuqing, T.; Bruton, C.J.; Chater, K.F. λ Red-Mediated Genetic Manipulation of Antibiotic-Producing Streptomyces. Elsevier 2004, 54, 107–128. [Google Scholar]
- Myronovskyi, M.; Welle, E.; Fedorenko, V.; Luzhetskyy, A. β-Glucuronidase as a Sensitive and Versatile Reporter in Actinomycetes. Appl. Environ. Microbiol. 2011, 77, 5370–5383. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tanaka, A.; Takano, Y.; Ohnishi, Y.; Horinouchi, S. AfsR Recruits RNA Polymerase to the afsS Promoter: A Model for Transcriptional Activation by SARPs. J. Mol. Biol. 2007, 369, 322–333. [Google Scholar] [CrossRef] [PubMed]
- Xiong, A.; Gottman, A.; Park, C.; Baetens, M.; Pandza, S.; Matin, A. The EmrR Protein Represses the Escherichia coli emrRAB Multidrug Resistance Operon by Directly Binding to Its Promoter Region. Antimicrob. Agents Chemother. 2000, 44, 2905–2907. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davis, J.R.; Sello, J.K. Regulation of genes in Streptomyces bacteria required for catabolism of lignin-derived aromatic compounds. Appl. Microbiol. Biotechnol. 2009, 86, 921–929. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.-M.; Jeng, W.-Y.; Ko, T.-P.; Yeh, Y.-J.; Chen, C.K.-M.; Wang, A.H.-J. Structural study of TcaR and its complexes with multiple antibiotics from Staphylococcus epidermidis. Proc. Natl. Acad. Sci. USA 2010, 107, 8617–8622. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fuangthong, M.; Atichartpongkul, S.; Mongkolsuk, S.; Helmann, J.D. OhrR Is a Repressor of ohrA, a Key Organic Hydroperoxide Resistance Determinant in Bacillus subtilis. J. Bacteriol. 2001, 183, 4134–4141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ellison, D.W.; Miller, V.L. Regulation of virulence by members of the MarR/SlyA family. Curr. Opin. Microbiol. 2006, 9, 153–159. [Google Scholar] [CrossRef]
- Perera, I.C.; Grove, A. Molecular Mechanisms of Ligand-Mediated Attenuation of DNA Binding by MarR Family Transcriptional Regulators. J. Mol. Cell Biol. 2010, 2, 243–254. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mast, Y.; Guezguez, J.; Handel, F.; Schinko, E. A Complex Signaling Cascade Governs Pristinamycin Biosynthesis in Streptomyces pristinaespiralis. Appl. Environ. Microbiol. 2015, 81, 6621–6636. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Handel, F.; Kulik, A.; Mast, Y. Investigation of the Autoregulator-Receptor System in the Pristinamycin Producer Streptomyces pristinaespiralis. Front. Microbiol. 2020, 11, 580990. [Google Scholar] [CrossRef]
- Wang, L.; Tian, X.; Wang, J.; Yang, H.; Fan, K.; Xu, G.; Yang, K.; Tan, H. Autoregulation of antibiotic biosynthesis by binding of the end product to an atypical response regulator. Proc. Natl. Acad. Sci. USA 2009, 106, 8617–8622. [Google Scholar] [CrossRef] [Green Version]
- Lee, P.-C.; Umeyama, T.; Horinouchi, S. afsS is a target of AfsR, a transcriptional factor with ATPase activity that globally controls secondary metabolism in Streptomyces coelicolor A3(2). Mol. Microbiol. 2002, 43, 1413–1430. [Google Scholar] [CrossRef]
- Horinouchi, S. AfsR as an integrator of signals that are sensed by multiple serine/threonine kinases in Streptomyces coelicolor A3(2). J. Ind. Microbiol. Biotechnol. 2003, 30, 462–467. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tsypik, O.; Makitrynskyy, R.; Yan, X.; Koch, H.-G.; Paululat, T.; Bechthold, A. Regulatory Control of Rishirilide(s) Biosynthesis in Streptomyces bottropensis. Microorganisms 2021, 9, 374. https://doi.org/10.3390/microorganisms9020374
Tsypik O, Makitrynskyy R, Yan X, Koch H-G, Paululat T, Bechthold A. Regulatory Control of Rishirilide(s) Biosynthesis in Streptomyces bottropensis. Microorganisms. 2021; 9(2):374. https://doi.org/10.3390/microorganisms9020374
Chicago/Turabian StyleTsypik, Olga, Roman Makitrynskyy, Xiaohui Yan, Hans-Georg Koch, Thomas Paululat, and Andreas Bechthold. 2021. "Regulatory Control of Rishirilide(s) Biosynthesis in Streptomyces bottropensis" Microorganisms 9, no. 2: 374. https://doi.org/10.3390/microorganisms9020374
APA StyleTsypik, O., Makitrynskyy, R., Yan, X., Koch, H.-G., Paululat, T., & Bechthold, A. (2021). Regulatory Control of Rishirilide(s) Biosynthesis in Streptomyces bottropensis. Microorganisms, 9(2), 374. https://doi.org/10.3390/microorganisms9020374





