Hypo- and Hyper-Virulent Listeria monocytogenes Clones Persisting in Two Different Food Processing Plants of Central Italy
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains
2.2. Molecular Serogrouping by PCR
2.3. Whole Genome Sequencing
2.3.1. In Silico Multi Locus Sequence Typing (MLST)
2.3.2. Core Genome MLST
2.3.3. Single Nucleotide Polymorphism (SNP) Analysis
2.3.4. Detection of Genetic Determinants Involved in Persistence
2.3.5. Virulence-Associated Genes
Statistical Analysis
2.4. Biofilm-Forming Ability In Vitro Assay
Statistical Analysis
3. Results
3.1. Whole Genome Sequencing
3.1.1. Meat A
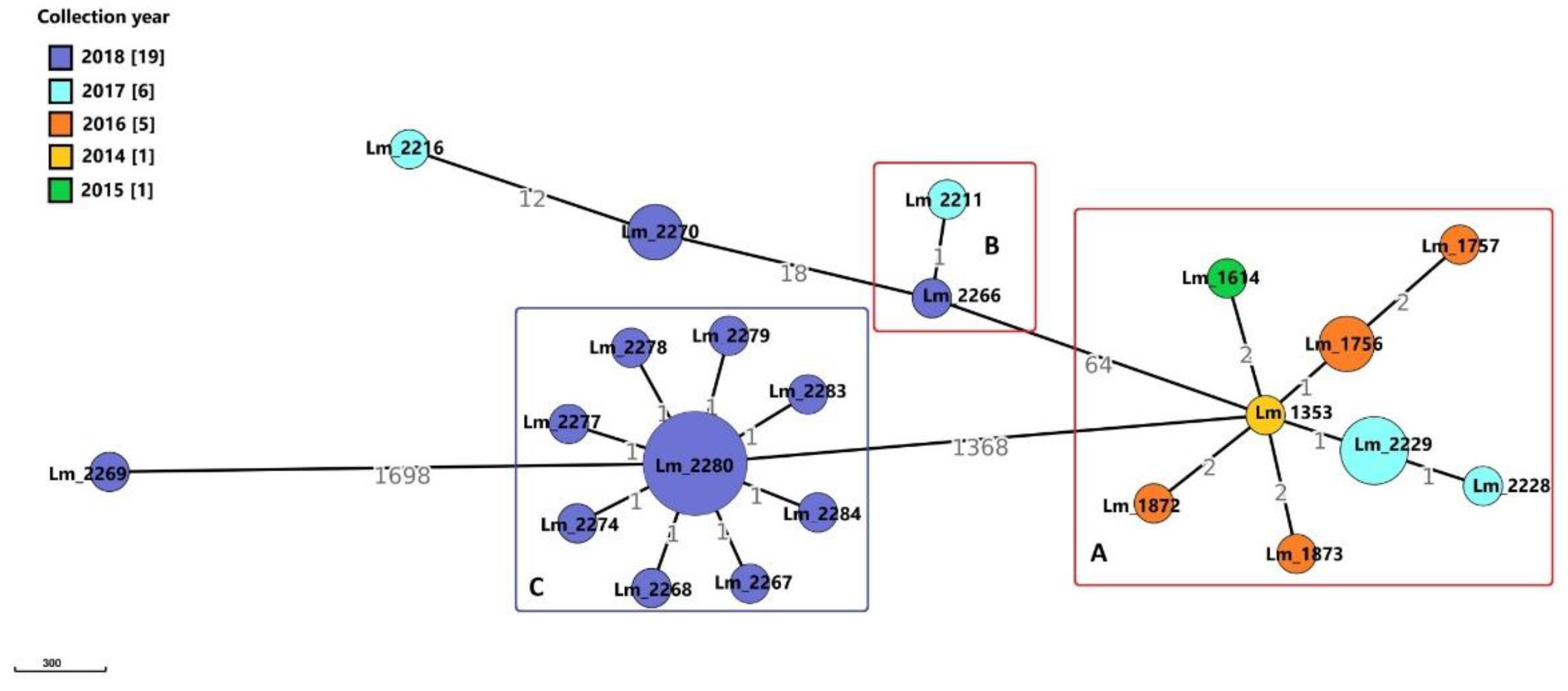
Molecular Typing and Cluster Analysis
Genetic Determinants Involved in Persistence
Virulence-Associated Genes
3.1.2. Dairy B
Molecular Typing and Cluster Analysis
Genetic Determinants Involved in Persistence
Virulence-Associated Genes
3.2. Biofilm-Forming Ability
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- European Food Safety Authority and European Centre for Disease Prevention and Control (EFSA and ECDC). The European Union One Health 2018 Zoonoses Report. EFSA J. 2019, 17. [Google Scholar] [CrossRef] [Green Version]
- Camargo, A.C.; Moura, A.; Avillan, J.; Herman, N.; McFarland, A.P.; Sreevatsan, S.; Call, D.R.; Woodward, J.J.; Lecuit, M.; Nero, L.A. Whole-genome sequencing reveals Listeria monocytogenes diversity and allows identification of long-term persistent strains in Brazil. Environ. Microbiol. 2019, 21, 4478–4487. [Google Scholar] [CrossRef]
- Stoller, A.; Stevens, M.; Stephan, R.; Guldimann, C. Characteristics of Listeria monocytogenes Strains Persisting in a Meat Processing Facility over a 4-Year Period. Pathogens 2019, 8, 32. [Google Scholar] [CrossRef] [Green Version]
- Maury, M.M.; Bracq-Dieye, H.; Huang, L.; Vales, G.; Lavina, M.; Thouvenot, P.; Disson, O.; Leclercq, A.; Brisse, S.; Lecuit, M. Hypervirulent Listeria monocytogenes clones’ adaption to mammalian gut accounts for their association with dairy products. Nat. Commun. 2019, 10, 2488. [Google Scholar] [CrossRef] [Green Version]
- Ratani, S.S.; Siletzky, R.M.; Dutta, V.; Yildirim, S.; Osborne, J.A.; Lin, W.; Hitchins, A.D.; Ward, T.J.; Kathariou, S. Heavy Metal and Disinfectant Resistance of Listeria monocytogenes from Foods and Food Processing Plants. Appl. Environ. Microbiol. 2012, 78, 6938. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pombinho, R.; Camejo, A.; Vieira, A.; Reis, O.; Carvalho, F.; Almeida, M.T.; Pinheiro, J.C.; Sousa, S.; Cabanes, D. Listeria monocytogenes CadC Regulates Cadmium Efflux and Fine-tunes Lipoprotein Localization to Escape the Host Immune Response and Promote Infection. J. Infect. Dis. 2017, 215, 1468–1479. [Google Scholar] [CrossRef] [PubMed]
- Mullapudi, S.; Siletzky, R.M.; Kathariou, S. Heavy-Metal and Benzalkonium Chloride Resistance of Listeria monocytogenes Isolates from the Environment of Turkey-Processing Plants. Appl. Environ. Microbiol. 2008, 74, 1464–1468. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.; Rakic-Martinez, M.; Graves, L.M.; Ward, T.J.; Siletzky, R.M.; Kathariou, S. Genetic Determinants for Cadmium and Arsenic Resistance among Listeria monocytogenes Serotype 4b Isolates from Sporadic Human Listeriosis Patients. Appl. Environ. Microbiol. 2013, 79, 2471–2476. [Google Scholar] [CrossRef] [Green Version]
- Parsons, C.; Lee, S.; Kathariou, S. Dissemination and conservation of cadmium and arsenic resistance determinants in Listeria and other Gram-positive bacteria. Mol. Microbiol. 2020, 113, 560–569. [Google Scholar] [CrossRef] [Green Version]
- Ordoñez, O.F.; Lanzarotti, E.; Kurth, D.; Cortez, N.; Farías, M.E.; Turjanski, A.G. Genome comparison of two Exiguobacterium strains from high altitude andean lakes with different arsenic resistance: Identification and 3D modeling of the Acr3 efflux pump. Front. Environ. Sci. 2015, 3. [Google Scholar] [CrossRef] [Green Version]
- Parsons, C.; Lee, S.; Kathariou, S. Heavy Metal Resistance Determinants of the Foodborne Pathogen Listeria monocytogenes. Genes 2018, 10, 11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuenne, C.; Billion, A.; Mraheil, M.A.; Strittmatter, A.; Daniel, R.; Goesmann, A.; Barbuddhe, S.; Hain, T.; Chakraborty, T. Reassessment of the Listeria monocytogenes pan-genome reveals dynamic integration hotspots and mobile genetic elements as major components of the accessory genome. BMC Genom. 2013, 14, 47. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Corbett, D.; Schuler, S.; Glenn, S.; Andrew, P.W.; Cavet, J.S.; Roberts, I.S. The combined actions of the copper-responsive repressor CsoR and copper-metallochaperone CopZ modulate CopA-mediated copper efflux in the intracellular pathogen Listeria monocytogenes: Copper homeostasis in Listeria monocytogenes. Mol. Microbiol. 2011, 81, 457–472. [Google Scholar] [CrossRef] [PubMed]
- Ciccio, P.D.; Chiesa, F.; Rubiola, S.; Civera, T. Genetic Determinants Associated with Biofilm Formation of Listeria monocytogenes from Food and Food Processing Environment. In Proceedings of the 33rd EFFoST International Conference Sustainable Food Systems-Performing by Connecting, Rotterdam, The Netherlands, 12–14 November 2019. [Google Scholar]
- Franciosa, G.; Maugliani, A.; Scalfaro, C.; Floridi, F.; Aureli, P. Expression of Internalin a and Biofilm Formation among Listeria monocytogenes Clinical Isolates. Int. J. Immunopathol. Pharmacol. 2009, 22, 183–193. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Conficoni, D.; Losasso, C.; Cortini, E.; Di Cesare, A.; Cibin, V.; Giaccone, V.; Corno, G.; Ricci, A. Resistance to Biocides in Listeria monocytogenes Collected in Meat-Processing Environments. Front. Microbiol. 2016, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tezel, U.; Pavlostathis, S.G. Quaternary ammonium disinfectants: Microbial adaptation, degradation and ecology. Curr. Opin. Biotechnol. 2015, 33, 296–304. [Google Scholar] [CrossRef]
- Cherifi, T.; Carrillo, C.; Lambert, D.; Miniaï, I.; Quessy, S.; Larivière-Gauthier, G.; Blais, B.; Fravalo, P. Genomic characterization of Listeria monocytogenes isolates reveals that their persistence in a pig slaughterhouse is linked to the presence of benzalkonium chloride resistance genes. BMC Microbiol. 2018, 18, 220. [Google Scholar] [CrossRef]
- Zuber, I.; Lakicevic, B.; Pietzka, A.; Milanov, D.; Djordjevic, V.; Karabasil, N.; Teodorovic, V.; Ruppitsch, W.; Dimitrijevic, M. Molecular characterization of Listeria monocytogenes isolates from a small-scale meat processor in Montenegro, 2011–2014. Food Microbiol. 2019, 79, 116–122. [Google Scholar] [CrossRef]
- Moura, A.; Tourdjman, M.; Leclercq, A.; Hamelin, E.; Laurent, E.; Fredriksen, N.; Van Cauteren, D.; Bracq-Dieye, H.; Thouvenot, P.; Vales, G.; et al. Real-Time Whole-Genome Sequencing for Surveillance of Listeria monocytogenes, France. Emerg. Infect. Dis. 2017, 23, 1462–1470. [Google Scholar] [CrossRef] [Green Version]
- Duranti, A.; Sabbatucci, M.; Blasi, G.; Acciari, V.A.; Ancora, M.; Bella, A.; Busani, L.; Centorame, P.; Cammà, C.; Conti, F.; et al. A severe outbreak of listeriosis in central Italy with a rare pulsotype associated with processed pork products. J. Med. Microbiol. 2018, 67, 1351–1360. [Google Scholar] [CrossRef]
- Torresi, M.; Ruolo, A.; Acciari, V.A.; Ancora, M.; Blasi, G.; Cammà, C.; Centorame, P.; Centorotola, G.; Curini, V.; Guidi, F.; et al. A Real-Time PCR Screening Assay for Rapid Detection of Listeria monocytogenes Outbreak Strains. Foods 2020, 9, 67. [Google Scholar] [CrossRef] [Green Version]
- Doumith, M.; Buchrieser, C.; Glaser, P.; Jacquet, C.; Martin, P. Differentiation of the Major Listeria monocytogenes Serovars by Multiplex PCR. J. Clin. Microbiol. 2004, 42, 3819–3822. [Google Scholar] [CrossRef] [Green Version]
- Kérouanton, A.; Marault, M.; Petit, L.; Grout, J.; Dao, T.T.; Brisabois, A. Evaluation of a multiplex PCR assay as an alternative method for Listeria monocytogenes serotyping. J. Microbiol. Methods 2010, 80, 134–137. [Google Scholar] [CrossRef] [PubMed]
- Cito, F.; Di Pasquale, A.; Cammà, C.; Cito, P. The Italian information system for the collection and analysis of complete genome sequence of pathogens isolated from animal, food and environment. Int. J. Infect. Dis. 2018, 73, 296–297. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [Green Version]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality assessment tool for genome assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef]
- Salcedo, C.; Arreaza, L.; Alcala, B.; de la Fuente, L.; Vazquez, J.A. Development of a Multilocus Sequence Typing Method for Analysis of Listeria monocytogenes Clones. J. Clin. Microbiol. 2003, 41, 757–762. [Google Scholar] [CrossRef] [Green Version]
- Silva, M.; Machado, M.P.; Silva, D.N.; Rossi, M.; Moran-Gilad, J.; Santos, S.; Ramirez, M.; Carriço, J.A. chewBBACA: A complete suite for gene-by-gene schema creation and strain identification. Microb. Genom. 2018, 4. [Google Scholar] [CrossRef]
- Zhou, Z.; Alikhan, N.-F.; Sergeant, M.J.; Luhmann, N.; Vaz, C.; Francisco, A.P.; Carriço, J.A.; Achtman, M. GrapeTree: Visualization of core genomic relationships among 100,000 bacterial pathogens. Genome Res. 2018, 28, 1395–1404. [Google Scholar] [CrossRef] [Green Version]
- Gardner, S.N.; Slezak, T.; Hall, B.G. kSNP3.0: SNP detection and phylogenetic analysis of genomes without genome alignment or reference genome: Table 1. Bioinformatics 2015, 31, 2877–2878. [Google Scholar] [CrossRef] [Green Version]
- Morganti, M.; Scaltriti, E.; Cozzolino, P.; Bolzoni, L.; Casadei, G.; Pierantoni, M.; Foni, E.; Pongolini, S. Processing-Dependent and Clonal Contamination Patterns of Listeria monocytogenes in the Cured Ham Food Chain Revealed by Genetic Analysis. Appl. Environ. Microbiol. 2016, 82, 822. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Fagerlund, A.; Langsrud, S.; Møretrø, T. In-Depth Longitudinal Study of Listeria monocytogenes ST9 Isolates from the Meat Processing Industry: Resolving Diversity and Transmission Patterns Using Whole-Genome Sequencing. Appl. Environ. Microbiol. 2020, 86, e00579-20. [Google Scholar] [CrossRef]
- Gerner-Smidt, P.; Besser, J.; Concepción-Acevedo, J.; Folster, J.P.; Huffman, J.; Joseph, L.A.; Kucerova, Z.; Nichols, M.C.; Schwensohn, C.A.; Tolar, B. Whole Genome Sequencing: Bridging One-Health Surveillance of Foodborne Diseases. Front. Public Health 2019, 7, 172. [Google Scholar] [CrossRef]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Carattoli, A.; Zankari, E.; García-Fernández, A.; Voldby Larsen, M.; Lund, O.; Villa, L.; Møller Aarestrup, F.; Hasman, H. In Silico Detection and Typing of Plasmids using PlasmidFinder and Plasmid Multilocus Sequence Typing. Antimicrob. Agents Chemother. 2014, 58, 3895–3903. [Google Scholar] [CrossRef] [Green Version]
- Seemann, T. Abricate—Mass Screening of Contigs for Antimicrobial and Virulence Genes. Available online: https://github.com/tseemann/abricate (accessed on 23 October 2020).
- Chen, L.; Zheng, D.; Liu, B.; Yang, J.; Jin, Q. VFDB 2016: Hierarchical and refined dataset for big data analysis—10 years on. Nucleic Acids Res. 2016, 44, D694–D697. [Google Scholar] [CrossRef]
- Alcock, B.P.; Raphenya, A.R.; Lau, T.T.Y.; Tsang, K.K.; Bouchard, M.; Edalatmand, A.; Huynh, W.; Nguyen, A.-L.V.; Cheng, A.A.; Liu, S.; et al. CARD 2020: Antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 2019, gkz935. [Google Scholar] [CrossRef]
- Bortolaia, V.; Kaas, R.S.; Ruppe, E.; Roberts, M.C.; Schwarz, S.; Cattoir, V.; Philippon, A.; Allesoe, R.L.; Rebelo, A.R.; Florensa, A.F.; et al. ResFinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 2020, 75, 3491–3500. [Google Scholar] [CrossRef]
- Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2020; Available online: https://www.R-project.org (accessed on 7 December 2020).
- Di Bonaventura, G.; Piccolomini, R.; Paludi, D.; D’Orio, V.; Vergara, A.; Conter, M.; Ianieri, A. Influence of temperature on biofilm formation by Listeria monocytogenes on various food-contact surfaces: Relationship with motility and cell surface hydrophobicity. J. Appl. Microbiol. 2008, 104, 1552–1561. [Google Scholar] [CrossRef]
- Canchaya, C.; Giubellini, V.; Ventura, M.; de los Reyes-Gavilán, C.G.; Margolles, A. Mosaic-Like Sequences Containing Transposon, Phage, and Plasmid Elements among Listeria monocytogenes Plasmids. Appl. Environ. Microbiol. 2010, 76, 4851–4857. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Zhou, Y.; Bao, H.; Zhang, L.; Wang, R.; Zhou, X. Plasmid-borne cadmium resistant determinants are associated with the susceptibility of Listeria monocytogenes to bacteriophage. Microbiol. Res. 2015, 172, 1–6. [Google Scholar] [CrossRef]
- Martín, B.; Perich, A.; Gómez, D.; Yangüela, J.; Rodríguez, A.; Garriga, M.; Aymerich, T. Diversity and distribution of Listeria monocytogenes in meat processing plants. Food Microbiol. 2014, 44, 119–127. [Google Scholar] [CrossRef]
- Rychli, K.; Stessl, B.; Szakmary-Brändle, K.; Strauß, A.; Wagner, M.; Schoder, D. Listeria monocytogenes Isolated from Illegally Imported Food Products into the European Union Harbor Different Virulence Factor Variants. Genes 2018, 9, 428. [Google Scholar] [CrossRef] [Green Version]
- EFSA Panel on Biological Hazards (BIOHAZ); Ricci, A.; Allende, A.; Bolton, D.; Chemaly, M.; Davies, R.; Fernández Escámez, P.S.; Girones, R.; Herman, L.; Koutsoumanis, K.; et al. Listeria monocytogenes contamination of ready-to-eat foods and the risk for human health in the EU. EFSA J. 2018, 16. [Google Scholar] [CrossRef]
- Félix, B.; Feurer, C.; Maillet, A.; Guillier, L.; Boscher, E.; Kerouanton, A.; Denis, M.; Roussel, S. Population Genetic Structure of Listeria monocytogenes Strains Isolated From the Pig and Pork Production Chain in France. Front. Microbiol. 2018, 9, 684. [Google Scholar] [CrossRef] [Green Version]
- Cantinelli, T.; Chenal-Francisque, V.; Diancourt, L.; Frezal, L.; Leclercq, A.; Wirth, T.; Lecuit, M.; Brisse, S. “Epidemic Clones” of Listeria monocytogenes Are Widespread and Ancient Clonal Groups. J. Clin. Microbiol. 2013, 51, 3770–3779. [Google Scholar] [CrossRef] [Green Version]
- Jiang, X.; Yu, T.; Liu, L.; Li, Y.; Zhang, K.; Wang, H.; Shi, L. Examination of Quaternary Ammonium Compound Resistance in Proteus mirabilis Isolated from Cooked Meat Products in China. Front. Microbiol. 2017, 8, 2417. [Google Scholar] [CrossRef]
- Tamburro, M.; Ripabelli, G.; Vitullo, M.; Dallman, T.J.; Pontello, M.; Amar, C.F.L.; Sammarco, M.L. Gene expression in Listeria monocytogenes exposed to sublethal concentration of benzalkonium chloride. Comp. Immunol. Microbiol. Infect. Dis. 2015, 40, 31–39. [Google Scholar] [CrossRef]
- Jiang, X.; Yu, T.; Xu, Y.; Wang, H.; Korkeala, H.; Shi, L. MdrL, a major facilitator superfamily efflux pump of Listeria monocytogenes involved in tolerance to benzalkonium chloride. Appl. Microbiol. Biotechnol. 2019, 103, 1339–1350. [Google Scholar] [CrossRef]
- Rakic-Martinez, M.; Drevets, D.A.; Dutta, V.; Katic, V.; Kathariou, S. Listeria monocytogenes Strains Selected on Ciprofloxacin or the Disinfectant Benzalkonium Chloride Exhibit Reduced Susceptibility to Ciprofloxacin, Gentamicin, Benzalkonium Chloride, and Other Toxic Compounds. Appl. Environ. Microbiol. 2011, 77, 8714–8721. [Google Scholar] [CrossRef] [Green Version]
- Hegstad, K.; Langsrud, S.; Lunestad, B.T.; Scheie, A.A.; Sunde, M.; Yazdankhah, S.P. Does the Wide Use of Quaternary Ammonium Compounds Enhance the Selection and Spread of Antimicrobial Resistance and Thus Threaten Our Health? Microb. Drug Resist. 2010, 16, 91–104. [Google Scholar] [CrossRef]
- Martínez-Suárez, J.V.; Ortiz, S.; López-Alonso, V. Potential Impact of the Resistance to Quaternary Ammonium Disinfectants on the Persistence of Listeria monocytogenes in Food Processing Environments. Front. Microbiol. 2016, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gadea, R.; Fernández Fuentes, M.Á.; Pérez Pulido, R.; Gálvez, A.; Ortega, E. Effects of exposure to quaternary-ammonium-based biocides on antimicrobial susceptibility and tolerance to physical stresses in bacteria from organic foods. Food Microbiol. 2017, 63, 58–71. [Google Scholar] [CrossRef] [PubMed]
- Harter, E.; Wagner, E.M.; Zaiser, A.; Halecker, S.; Wagner, M.; Rychli, K. Stress Survival Islet 2, Predominantly Present in Listeria monocytogenes Strains of Sequence Type 121, Is Involved in the Alkaline and Oxidative Stress Responses. Appl. Environ. Microbiol. 2017, 83, e00827-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ryan, S.; Begley, M.; Hill, C.; Gahan, C.G.M. A five-gene stress survival islet (SSI-1) that contributes to the growth of Listeria monocytogenes in suboptimal conditions: Stress survival islet in L. monocytogenes. J. Appl. Microbiol. 2010, 109, 984–995. [Google Scholar] [CrossRef] [PubMed]
- Keeney, K.; Trmcic, A.; Zhu, Z.; Delaquis, P.; Wang, S. Stress survival islet 1 contributes to serotype-specific differences in biofilm formation in Listeria monocytogenes. Lett. Appl. Microbiol. 2018, 67, 530–536. [Google Scholar] [CrossRef]
- Harrand, A.S.; Jagadeesan, B.; Baert, L.; Wiedmann, M.; Orsi, R.H. Evolution of Listeria monocytogenes in a Food Processing Plant Involves Limited Single-Nucleotide Substitutions but Considerable Diversification by Gain and Loss of Prophages. Appl. Environ. Microbiol. 2020, 86, e02493-19. [Google Scholar] [CrossRef] [PubMed]
- Parsons, C.; Lee, S.; Jayeola, V.; Kathariou, S. Novel Cadmium Resistance Determinant in Listeria monocytogenes. Appl. Environ. Microbiol. 2017, 83, e02580-16. [Google Scholar] [CrossRef] [Green Version]
- Azeredo, J.; Azevedo, N.F.; Briandet, R.; Cerca, N.; Coenye, T.; Costa, A.R.; Desvaux, M.; Di Bonaventura, G.; Hébraud, M.; Jaglic, Z.; et al. Critical review on biofilm methods. Crit. Rev. Microbiol. 2017, 43, 313–351. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ortiz, S.; López-Alonso, V.; Rodríguez, P.; Martínez-Suárez, J.V. The Connection between Persistent, Disinfectant-Resistant Listeria monocytogenes Strains from Two Geographically Separate Iberian Pork Processing Plants: Evidence from Comparative Genome Analysis. Appl. Environ. Microbiol. 2016, 82, 308–317. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pasquali, F.; Palma, F.; Guillier, L.; Lucchi, A.; De Cesare, A.; Manfreda, G. Listeria monocytogenes Sequence Types 121 and 14 Repeatedly Isolated Within One Year of Sampling in a Rabbit Meat Processing Plant: Persistence and Ecophysiology. Front. Microbiol. 2018, 9, 596. [Google Scholar] [CrossRef] [PubMed]
- Tavares, R.d.M.; da Silva, D.A.L.; Camargo, A.C.; Yamatogi, R.S.; Nero, L.A. Interference of the acid stress on the expression of llsX by Listeria monocytogenes pathogenic island 3 (LIPI-3) variants. Food Res. Int. 2020, 132, 109063. [Google Scholar] [CrossRef] [PubMed]
- Cotter, P.D.; Draper, L.A.; Lawton, E.M.; Daly, K.M.; Groeger, D.S.; Casey, P.G.; Ross, R.P.; Hill, C. Listeriolysin S, a Novel Peptide Haemolysin Associated with a Subset of Lineage I Listeria monocytogenes. PLoS Pathog. 2008, 4, e1000144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vilchis-Rangel, R.E.; Espinoza-Mellado, M.d.R.; Salinas-Jaramillo, I.J.; Martinez-Peña, M.D.; Rodas-Suárez, O.R. Association of Listeria monocytogenes LIPI-1 and LIPI-3 marker llsX with invasiveness. Curr. Microbiol. 2019, 76, 637–643. [Google Scholar] [CrossRef] [PubMed]
- Su, X.; Cao, G.; Zhang, J.; Pan, H.; Zhang, D.; Kuang, D.; Yang, X.; Xu, X.; Shi, X.; Meng, J. Characterization of internalin genes in Listeria monocytogenes from food and humans, and their association with the invasion of Caco-2 cells. Gut Pathog. 2019, 11, 30. [Google Scholar] [CrossRef] [PubMed]







| Genetic Determinant Category | Gene or Islet | Specific Location | Cluster or Isolate | Predicted Resistance Functions | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Meat A | Dairy B | |||||||||
| Cluster A | Cluster B | Cluster C | Lm_2270-2272 | Lm_2216 | Lm_2269 | Cluster D | ||||
| SMR | sugE | + | + | + | + | + | + | + | Multidrug efflux-pumps | |
| MFS | Mdrl | + | + | + | + | + | + | + | ||
| Lde | + | + | + | + | + | + | + | |||
| MATE | norM | + | + | + | + | + | + | + | ||
| mepA | + | + | + | + | + | + | + | |||
| QAC-specific resistance genes | qacH | Tn6188 | - | - | + | - | - | - | - | QAC resistance |
| Heavy metals resistance genes | arsA | + | + | - | + | + | - | - | Arsenic resistance | |
| arsA1 | LGI2 | - | - | - | - | - | - | + | ||
| arsA2 | LGI2 | - | - | - | - | - | - | + | ||
| arsB | + | + | + | + | + | + | + | |||
| arsC | + | + | + | + | + | + | + | |||
| arsD | + | + | - | + | + | - | - | |||
| arsD1 | LGI2 | - | - | - | - | - | - | + | ||
| arsD2 | LGI2 | - | - | - | - | - | - | + | ||
| acr3 | - | |||||||||
| cadA1 | pLM33 | + | - | - | - | - | - | - | Cadmium resistance | |
| pLM5578 | - | - | + | - | - | - | - | |||
| cadA4 | LGI2 | - | - | - | - | - | - | + | ||
| cadC1 | pLM33 | + | - | - | - | - | - | - | ||
| pLM5578 | - | - | + | - | - | - | - | |||
| csoR | + | + | + | + | + | + | + | Copper resistance | ||
| copA | + | + | + | + | + | + | + | |||
| copZ | + | + | + | + | + | + | + | |||
| copY | + | - | - | - | - | - | - | |||
| copB | + | - | - | - | - | - | - | |||
| Stress survival determinants and Islet | SSI-1 | + | + | - | + | + | + | - | Tolerance to low pH, high osmolarity, bile and nisin | |
| SSI-2 | - | - | + | - | - | - | - | Alkaline and oxidative stress resistance | ||
| gbuA | + | + | + | + | + | + | + | Osmotic stress resistance | ||
| gbuB | + | + | + | + | + | + | + | |||
| gbuC | + | + | + | + | + | + | + | |||
| npr | pLM5578 | - | - | + | - | - | - | - | Oxidative stress resistance | |
| + | + | - | + | + | + | - | ||||
| Lm_1306 | Lm_1311 | Lm_1431 | Lm_1607 | Lm_1671 | Lm_1672 | Lm_1746 | Lm_1813 | |
|---|---|---|---|---|---|---|---|---|
| Lm_1306 | 1 | 0.001 | 0.338 | 0.586 | 0.039 | <0.0001 | 0.002 | 0.001 |
| Lm_1311 | 0.001 | 1 | 0.016 | 0.005 | 0.193 | 0.009 | 0.800 | 0.956 |
| Lm_1431 | 0.338 | 0.016 | 1 | 0.679 | 0.270 | <0.0001 | 0.031 | 0.019 |
| Lm_1607 | 0.586 | 0.005 | 0.679 | 1 | 0.129 | <0.0001 | 0.010 | 0.006 |
| Lm_1671 | 0.039 | 0.193 | 0.270 | 0.129 | 1 | <0.0001 | 0.294 | 0.212 |
| Lm_1672 | <0.0001 | 0.009 | <0.0001 | <0.0001 | <0.0001 | 1 | 0.004 | 0.007 |
| Lm_1746 | 0.002 | 0.800 | 0.031 | 0.010 | 0.294 | 0.004 | 1 | 0.844 |
| Lm_1813 | 0.001 | 0.956 | 0.019 | 0.006 | 0.212 | 0.007 | 0.844 | 1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guidi, F.; Orsini, M.; Chiaverini, A.; Torresi, M.; Centorame, P.; Acciari, V.A.; Salini, R.; Palombo, B.; Brandi, G.; Amagliani, G.; et al. Hypo- and Hyper-Virulent Listeria monocytogenes Clones Persisting in Two Different Food Processing Plants of Central Italy. Microorganisms 2021, 9, 376. https://doi.org/10.3390/microorganisms9020376
Guidi F, Orsini M, Chiaverini A, Torresi M, Centorame P, Acciari VA, Salini R, Palombo B, Brandi G, Amagliani G, et al. Hypo- and Hyper-Virulent Listeria monocytogenes Clones Persisting in Two Different Food Processing Plants of Central Italy. Microorganisms. 2021; 9(2):376. https://doi.org/10.3390/microorganisms9020376
Chicago/Turabian StyleGuidi, Fabrizia, Massimiliano Orsini, Alexandra Chiaverini, Marina Torresi, Patrizia Centorame, Vicdalia Aniela Acciari, Romolo Salini, Barbara Palombo, Giorgio Brandi, Giulia Amagliani, and et al. 2021. "Hypo- and Hyper-Virulent Listeria monocytogenes Clones Persisting in Two Different Food Processing Plants of Central Italy" Microorganisms 9, no. 2: 376. https://doi.org/10.3390/microorganisms9020376
APA StyleGuidi, F., Orsini, M., Chiaverini, A., Torresi, M., Centorame, P., Acciari, V. A., Salini, R., Palombo, B., Brandi, G., Amagliani, G., Schiavano, G. F., Massacci, F. R., Fisichella, S., Domenico, M. D., Ancora, M., Pasquale, A. D., Duranti, A., Cammà, C., Pomilio, F., & Blasi, G. (2021). Hypo- and Hyper-Virulent Listeria monocytogenes Clones Persisting in Two Different Food Processing Plants of Central Italy. Microorganisms, 9(2), 376. https://doi.org/10.3390/microorganisms9020376







