Presence of Chromosomal crpP-like Genes Is Not Always Associated with Ciprofloxacin Resistance in Pseudomonas aeruginosa Clinical Isolates Recovered in ICU Patients from Portugal and Spain
Abstract
1. Introduction
2. Materials and Methods
2.1. Pseudomonas aeruginosa Isolates and Antimicrobial Susceptibility
2.2. Sequence Analysis
2.3. Ciprofloxacin Resistance Determinants
2.4. Genetic Context of crpP Gene
2.5. Statistical Analysis
2.6. Accession Numbers
3. Results
3.1. Ciprofloxacin Resistance in STEP and SUPERIOR P. aeruginosa Isolates
3.2. P. aeruginosa Genome Analysis and Molecular Typing
3.3. Acquired Resistance Genes
3.4. Mutational Resistome
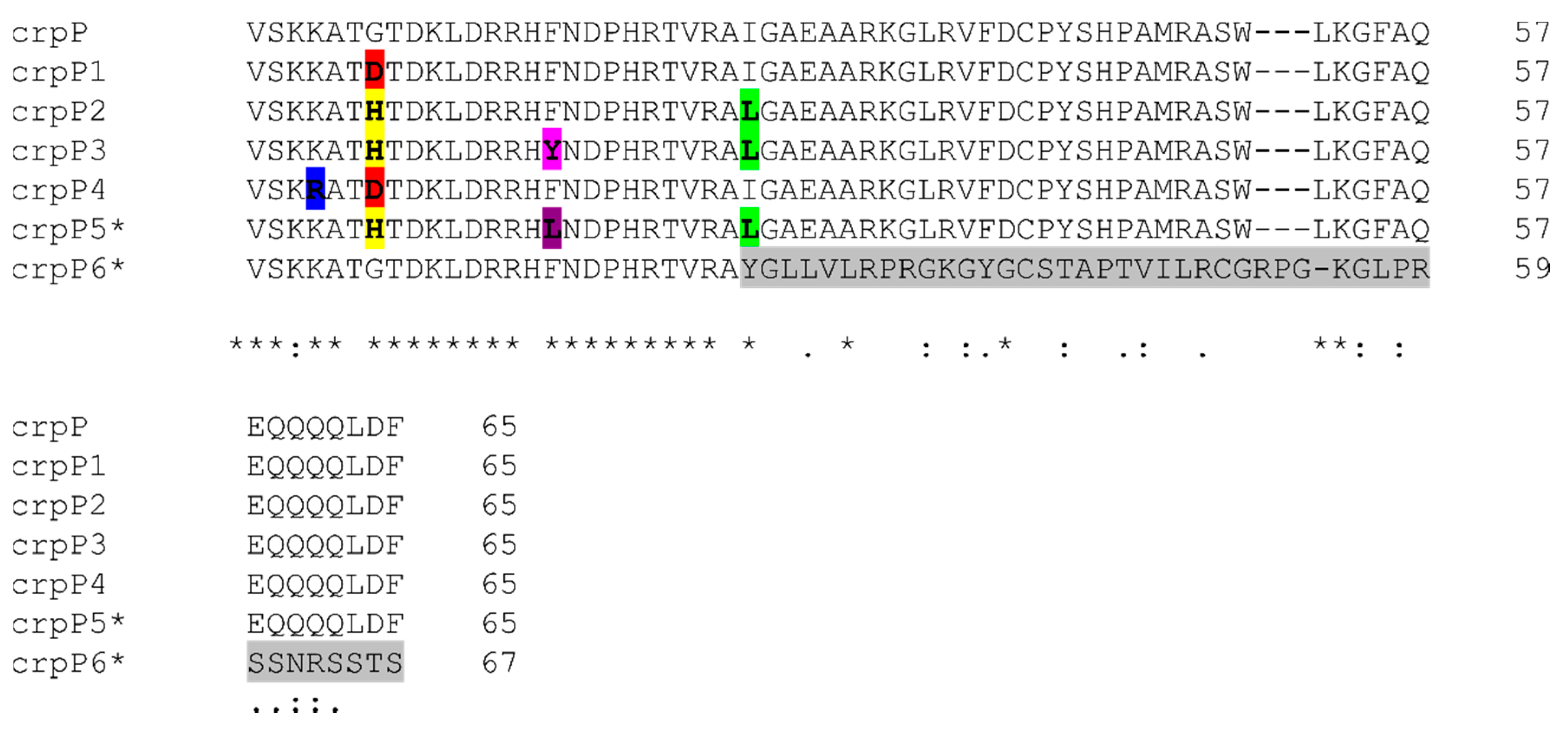
3.5. crpP-Encoding Genes
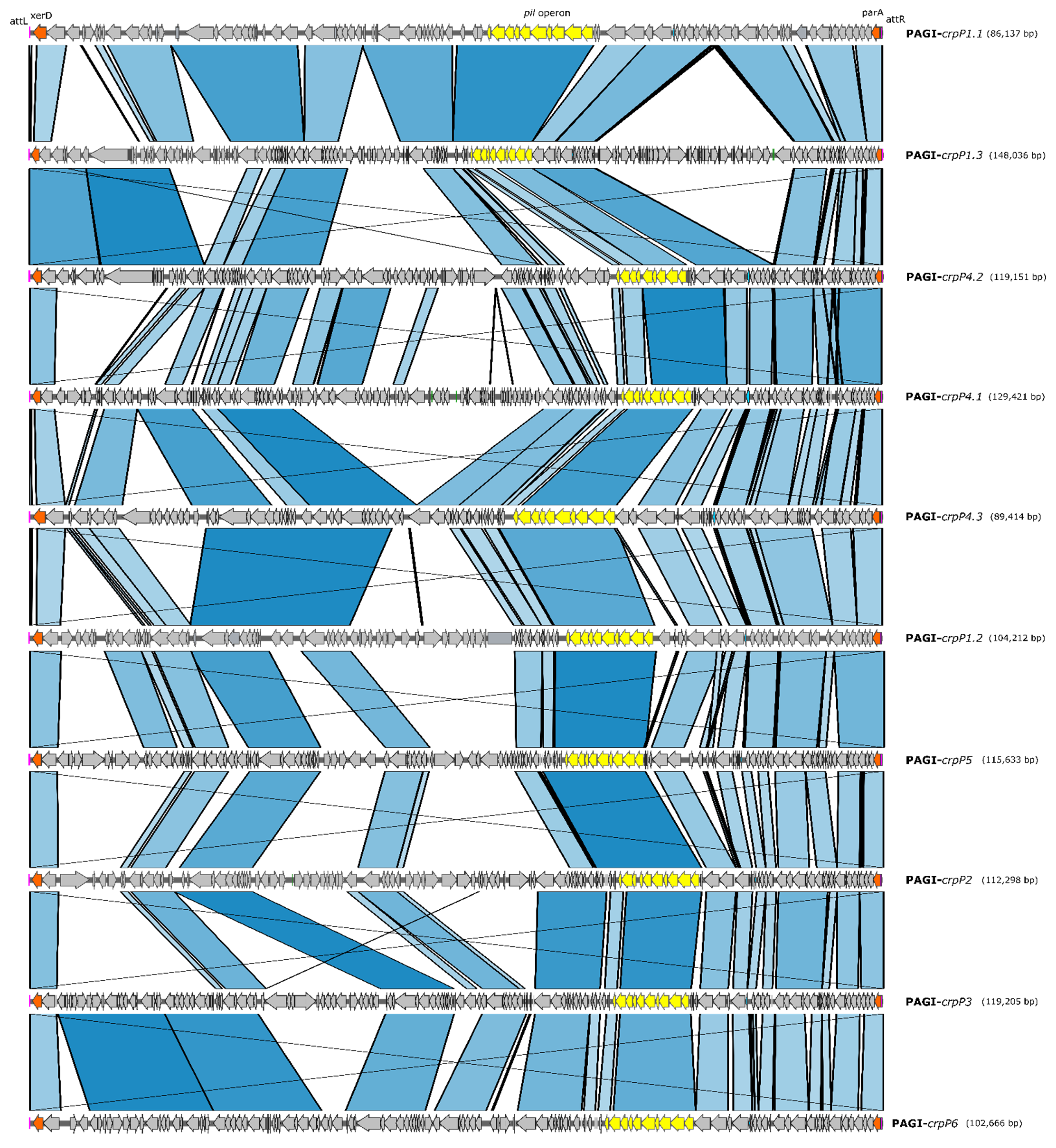
3.6. crpP-Carrying Pathogenicity Genomic Islands (PAGI)
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Magiorakos, A.P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef]
- Kadri, S.S.; Adjemian, J.; Lai, Y.L.; Spaulding, A.B.; Ricotta, E.; Rebecca Prevots, D.; Palmore, T.N.; Rhee, C.; Klompas, M.; Dekker, J.P.; et al. Difficult-to-treat resistance in gram-negative bacteremia at 173 US hospitals: Retrospective cohort analysis of prevalence, predictors, and outcome of resistance to all first-line agents. Clin. Infect. Dis. 2018, 67, 1803–1814. [Google Scholar] [CrossRef] [PubMed]
- Hooper, D.C.; Jacoby, G.A. Topoisomerase inhibitors: Fluoroquinolone mechanisms of action and resistance. Cold Spring Harb. Perspect. Med. 2016, 6, a025320. [Google Scholar] [CrossRef] [PubMed]
- Bush, N.G.; Diez-santos, I.; Abbott, L.R.; Maxwell, A. Quinolones: Mechanism, lethality and their contributions to antibiotic resistance. Molecules 2020, 25, 5662. [Google Scholar] [CrossRef] [PubMed]
- Ruiz, J.; Pons, M.J.; Gomes, C. Transferable mechanisms of quinolone resistance. Int. J. Antimicrob. Agents 2012, 40, 196–203. [Google Scholar] [CrossRef] [PubMed]
- Ruiz, J. Transferable mechanisms of quinolone resistance from 1998 onward. Clin. Microbiol. Rev. 2019, 32, 1–59. [Google Scholar] [CrossRef] [PubMed]
- Naeem, A.; Badshah, S.L.; Muska, M.; Ahmad, N.; Khan, K. The current case of quinolones: Synthetic approaches and antibacterial activity. Molecules 2016, 21, 268. [Google Scholar] [CrossRef] [PubMed]
- Rehman, A.; Patrick, W.M.; Lamont, I.L. Mechanisms of ciprofloxacin resistance in Pseudomonas aeruginosa: New approaches to an old problem. J. Med. Microbiol. 2019, 68, 1–10. [Google Scholar] [CrossRef]
- Chávez-jacobo, V.M.; Hernández-ramírez, K.C.; Romo-rodríguez, P.; Pérez-gallardo, R.V. CrpP is a novel ciprofloxacin-modifying enzyme encoded by the Pseudomonas aeruginosa pUM505 plasmid. Antimicrob. Agents Chemother. 2018, 62, 1–11. [Google Scholar] [CrossRef]
- Botelho, J.; Grosso, F.; Peixe, L. ICEs are the main reservoirs of the ciprofloxacin-modifying crpP Gene in Pseudomonas aeruginosa. Genes 2020, 11, 889. [Google Scholar] [CrossRef] [PubMed]
- Ortiz de la Rosa, J.M.; Nordmann, P.; Poirel, L. PAGI-associated CrpP-like fluoroquinolone-modifying enzymes among Pseudomonas aeruginosa clinical isolates in Europe. Antimicrob. Agents Chemother. 2020. [Google Scholar] [CrossRef]
- Chávez-Jacobo, V.M.; García Merinos, J.P.; López, Y.; Meza-Carmen, V.; Ramírez-Díaz, M.I. Identification of essential residues for ciprofloxacin resistance of ciprofloxacin-modifying enzyme (CrpP) of pUM505. Microbiology 2020, 367–374. [Google Scholar] [CrossRef]
- Chávez-Jacobo, V.; Hernández-Ramírez, K.C.; Silva-Sánchez, J.; Garza-Ramos, U.; Barrios-Camacho, H.; Ortiz-Alvarado, R.; Cervantes, C.; Meza-Carmen, V.; Ramírez-Díaz, M.I. Prevalence of the crpP gene conferring decreased ciprofloxacin susceptibility in enterobacterial clinical isolates from Mexican hospitals. J. Antimicrob. Chemother. 2019. [Google Scholar] [CrossRef]
- García-Fernández, S.; García-Castillo, M.; Melo-Cristino, J.; Pinto, M.F.; Gonçalves, E.; Alves, V.; Vieira, A.R.; Ramalheira, E.; Sancho, L.; Diogo, J.; et al. In vitro activity of ceftolozane-tazobactam against Enterobacterales and Pseudomonas aeruginosa causing urinary, intra-abdominal and lower respiratory tract infections in intensive care units in Portugal: The STEP multicenter study. Int. J. Antimicrob. Agents 2020. [Google Scholar] [CrossRef] [PubMed]
- García-Fernández, S.; García-Castillo, M.; Bou, G.; Calvo, J.; Cercenado, E.; Delgado, M.; Pitart, C.; Mulet, X.; Tormo, N.; Mendoza, D.L.; et al. Activity of ceftolozane/tazobactam against Pseudomonas aeruginosa and Enterobacterales isolates recovered from intensive care unit patients in Spain: The SUPERIOR multicentre study. Int. J. Antimicrob. Agents 2019, 53, 682–688. [Google Scholar] [CrossRef] [PubMed]
- Hernández-García, M.; García-Castillo, M.; García-Fernández, S.; Melo-Cristino, J.; Pinto, M.F.; Goncalves, E.; Alves, V.; Vieira, A.R.; Elmano, R.; Sancho, L.; et al. Distinct epidemiology and resistance mechanisms affecting ceftolozane/tazobactam in Pseudomonas aeruginosa isolates recovered from ICU patients in Spain and Portugal depicted by WGS. J. Antimicrob. Chemother. 2020, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Schmieder, R.; Edwards, R. Quality control and preprocessing of metagenomic datasets. Bioinformatics 2011, 27, 863–864. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.O.N.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality assessment tool for genome assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef]
- Wood, D.E.; Salzberg, S.L. Kraken: Ultrafast metagenomic sequence classification using exact alignments. Genome Biol. 2014, 15. [Google Scholar] [CrossRef]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Wintersinger, J.A.; Wasmuth, J.D. Kablammo: An interactive, web-based BLAST results visualizer. Bioinformatics 2015, 31, 1305–1306. [Google Scholar] [CrossRef]
- van Duin, D.; Bonomo, R.A. Ceftazidime/avibactam and ceftolozane/tazobactam: Second-generation β-lactam/β-lactamase inhibitor combinations. Clin. Infect. Dis. 2016, 63, 234–241. [Google Scholar] [CrossRef]
- Hernández-García, M.; García-fernández, S.; García-castillo, M.; Melo-Cristino, J.; Pinto, M.F.; Goncalves, E.; Alves, V.; Costa, E.; Ramalheira, E.; Sancho, L.; et al. Confronting ceftolozane-tazobactam susceptibility in multidrug-resistant Enterobacterales isolates and whole-genome sequencing results (STEP study). Int. J. Antimicrob. Agents 2020. [Google Scholar] [CrossRef]
- Hernández-García, M.; García-Fernández, S.; García-Castillo, M.; Bou, G.; Cercenado, E.; Dalgado-Valverde, M.; Mulet, X.; Pitart, C.; Rodríguez-Lozano, J.; Tormo, N.; et al. WGS characterization of MDR Enterobacterales with different ceftolozane/tazobactam susceptibility profiles during the SUPERIOR surveillance study in Spain. JAC Antimicrob. Resist. 2020. [Google Scholar] [CrossRef]
- Ruiz, J. CrpP, a passenger or a hidden stowaway in the Pseudomonas aeruginosa genome? J. Antimicrob. Chemother. 2019, 10–12. [Google Scholar] [CrossRef] [PubMed]
- Qiu, X.; Gurkar, A.U.; Lory, S. Interstrain transfer of the large pathogenicity island (PAPI-1) of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 2006, 103, 19830–19835. [Google Scholar] [CrossRef]
- Cabot, G.; López-Causapé, C.; Ocampo-Sosa, A.A.; Sommer, L.M.; Domínguez, M.Á.; Zamorano, L.; Juan, C.; Tubau, F.; Rodríguez, C.; Moyà, B.; et al. Deciphering the resistome of the widespread Pseudomonas sequence type 175 international high-risk clone through whole-genome sequencing. Antimicrob. Agents Chemother. 2016, 60, 7415–7423. [Google Scholar] [CrossRef] [PubMed]
- García-Castillo, M.; Del Campo, R.; Morosini, M.I.; Riera, E.; Cabot, G.; Willems, R.; Van Mansfeld, R.; Oliver, A.; Cantón, R. Wide dispersion of ST175 clone despite high genetic diversity of carbapenem-nonsusceptible Pseudomonas aeruginosa clinical strains in 16 Spanish hospitals. J. Clin. Microbiol. 2011, 49, 2905–2910. [Google Scholar] [CrossRef]
- Michael, B.M.F.; Lawrence, L.R. Brown genomic islands of Pseudomonas aeruginosa. FEMS Microbiol. Lett. 2009, 290. [Google Scholar] [CrossRef]
| CT-R | CT-S | p-Value | Odd Ratio (95%CI) | ||||
|---|---|---|---|---|---|---|---|
| Total No. | CIP-R No. (%) | CIP-S No. (%) | CIP-R No. (%) | CIP-S No. (%) | |||
| STEP | 396 | 19 (90.5) | 2 (9.5) | 128 (34.1) | 247 (65.9) | <0.001 | 18.2 (4.3–163.5) |
| SUPERIOR | 80 | 6 (85.7) | 1 (14.3) | 22 (30.1) | 51 (69.9) | 0.006 | 13.4 (1.5–649.5) |
| Total (n = 55) | CIP a Susceptibility Profile * | p-Value | Odd Ratio (95%CI) | ||
|---|---|---|---|---|---|
| I [No. (%)] (n = 9) | R [No. (%)] (n = 46) | ||||
| ARG b | 6 (10.9) | 0 | 6 (13) | 0.57 | 0 (0–4.55) |
| qnrS2 | 2 (3.6) | 0 | 2 (4.3) | 0.57 | 0 (0–4.55) |
| aac(6′)-Ib-cr | 6 (10.9) | 0 | 6 (13) | 1 | 0 (0–28.23) |
| QRDR c | 47 (85.5) | 5 (55.6) | 42 (91.3) | 0.02 | 7.90 (1.12–59.46) |
| GyrA | 42 (76.4) | 4 (44.4) | 38 (82.6) | 0.03 | 5.69 (0.99–35.95) |
| GyrB | 1 (1.8) | 0 | 1 (2.2) | NA | NA |
| ParC | 40 (72.7) | 3 (33.3) | 37 (80.4) | 0.01 | 7.81 (1.37–57.90) |
| ParE | 26 (47.3) | 4 (44.4) | 22 (47.8) | 1 | 1.14 (0.21–6.54) |
| CrpP d | 36 (65.5) | 8 (88.9) | 28 (60.9) | 0.14 | 0.20 (0.01–1.70) |
| CrpP1 | 23 (41.8) | 2 (22.2) | 21 (45.6) | 0.27 | 2.89 (0.48–31.41) |
| CrpP2 | 4 (7.2) | 3 (33.3) | 1 (2.2) | 0.01 | 0.05 (0.001–0.72) |
| CrpP3 | 4 (7.2) | 0 | 4 (8.7) | NA | NA |
| CrpP4 | 3 (5.4) | 1 (11.1) | 2 (4.3) | 0.07 | 11.89 (0.55–769.73) |
| CrpP5 | 1 (1.8) | 1 (11.1) | 0 | NA | NA |
| CrpP6 | 1 (1.8) | 0 | 1 (2.2) | NA | NA |
| nalCe | 48 (87.3) | 6 (66.7) | 42 (91.3) | 0.08 | 0.20 (0.03–1.70) |
| nfxBe | 4 (7.3) | 2 (22.2) | 2 (4.3) | 0.12 | 5.97 (0.38–95.35) |
| Enzyme (No. of Isolates) | Gene (No. of Isolates) | Mutations (nt) * | Missense Mutations (aa) * | Clonal Complex (No. of Isolates) | CIP-MIC Interpretation (No. of Isolates) |
|---|---|---|---|---|---|
| CrpP1 (23) | crpP1.1 (20) | 20(A-G), 192(C-T) | G7D | CC175 (10), CC348 (5), CC253 (3), CC179 (1), CC308 (1) | I (2), R (18) |
| crpP1.2 (2) | 20(A-G), 66(C-T) | CC554 (2) | R (2) | ||
| crpP1.3 (1) | 1 (T-C), 20(A-G), 192(C-T) | CC235 (1) | R (1) | ||
| CrpP2 (4) | crpP2 (4) | 19(CA-GG), 39(G-A), 45(T-C), 76(C-A), 81(C-T), 123(T-C), 183(A-G), 192(C-T) | G7H, I26L | CC244 (4) | I (3), R (1) |
| CrpP3 (4) | crpP3 (4) | 19(CA-GG), 39(G-A), 45(T-C), 47(A-T), 76(C-A), 81(C-T), 123(T-C), 183(A-G), 192(C-T) | G7H, F16Y, I26L | CC244 (3), CC313 (1) | I (1), R (3) |
| CrpP4 (3) | crpP4.1 (1) | 11(A-G), 20(A-G), 138(A-G) | K4R, G7D | CC27 (1) | I (1) |
| crpP4.2 (1) | 11(A-G), 20(A-G), 192(C-T) | CC446 (1) | R (1) | ||
| crpP4.3 (1) | 11(A-G), 18(T-C), 20(A-G), 174(A-G), 183 (A-G) | CC179 (1) | R (1) | ||
| CrpP5 (1) | crpP5 (1) | 19(CA-GG), 39(G-A), 45(CT-TC), 76(C-A), 81(C-T), 123(T-C), 162(A-G), 183(A-G), 192(C-T) | G7H, F16L, I26L | CC971 (1) | I (1) |
| CrpP6 (1) | crpP6 (1) | 33(A-G), 73(insGCTTACGG) | I26fs | CC499 (1) | R (1) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hernández-García, M.; García-Castillo, M.; García-Fernández, S.; López-Mendoza, D.; Díaz-Regañón, J.; Romano, J.; Pássaro, L.; Paixão, L.; Cantón, R. Presence of Chromosomal crpP-like Genes Is Not Always Associated with Ciprofloxacin Resistance in Pseudomonas aeruginosa Clinical Isolates Recovered in ICU Patients from Portugal and Spain. Microorganisms 2021, 9, 388. https://doi.org/10.3390/microorganisms9020388
Hernández-García M, García-Castillo M, García-Fernández S, López-Mendoza D, Díaz-Regañón J, Romano J, Pássaro L, Paixão L, Cantón R. Presence of Chromosomal crpP-like Genes Is Not Always Associated with Ciprofloxacin Resistance in Pseudomonas aeruginosa Clinical Isolates Recovered in ICU Patients from Portugal and Spain. Microorganisms. 2021; 9(2):388. https://doi.org/10.3390/microorganisms9020388
Chicago/Turabian StyleHernández-García, Marta, María García-Castillo, Sergio García-Fernández, Diego López-Mendoza, Jazmín Díaz-Regañón, João Romano, Leonor Pássaro, Laura Paixão, and Rafael Cantón. 2021. "Presence of Chromosomal crpP-like Genes Is Not Always Associated with Ciprofloxacin Resistance in Pseudomonas aeruginosa Clinical Isolates Recovered in ICU Patients from Portugal and Spain" Microorganisms 9, no. 2: 388. https://doi.org/10.3390/microorganisms9020388
APA StyleHernández-García, M., García-Castillo, M., García-Fernández, S., López-Mendoza, D., Díaz-Regañón, J., Romano, J., Pássaro, L., Paixão, L., & Cantón, R. (2021). Presence of Chromosomal crpP-like Genes Is Not Always Associated with Ciprofloxacin Resistance in Pseudomonas aeruginosa Clinical Isolates Recovered in ICU Patients from Portugal and Spain. Microorganisms, 9(2), 388. https://doi.org/10.3390/microorganisms9020388







