Prospection of Fungal Lignocellulolytic Enzymes Produced from Jatoba (Hymenaea courbaril) and Tamarind (Tamarindus indica) Seeds: Scaling for Bioreactor and Saccharification Profile of Sugarcane Bagasse
Abstract
:1. Introduction
2. Material and Methods
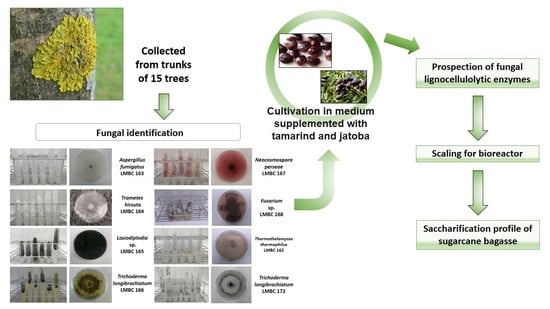
2.1. Sampling and Isolation of Filamentous Fungi
2.2. Plant Material
2.3. Fungal Identification
2.3.1. DNA Extraction
2.3.2. Polymerase Chain Reaction Amplification
2.3.3. DNA Sequencing
2.4. Selection of Microorganisms and Enzymatic Extract Preparations
2.5. Enzymatic Assays
2.5.1. Enzyme Determination Using Natural Substrates
2.5.2. Enzyme Determination with Synthetic Substrates
2.5.3. Scaling for Bioreactor
- Ni = stirring speed (1/s);
- tm = mixing time constant;
- V = volume of medium;
- Di = impeller diameter.
2.5.4. Protein Quantification
2.5.5. Pretreatment of Lignocellulosic Biomass
2.5.6. Enzymatic Hydrolysis and Determination of Sugars Content in Hydrolysates
2.6. Statistical Analysis
3. Results and Discussion
3.1. Identification of Microorganisms
3.2. Prospection of Microorganisms and Enzymatic Production
3.2.1. Time-Course of Enzyme Production from T. thermophilus LMBC 162: Culture with Jatoba Seeds in Agitated and Static Conditions
3.2.2. Time-Course of Enzyme Production from T. thermophilus LMBC 162: Culture with Tamarind Seeds in Agitated and Static Conditions
3.2.3. Time-Course of Enzyme Production from T. longibrachiatum LMBC 172: Culture with Jatoba Seeds in Agitated and Static Conditions
3.2.4. Time-Course of Enzyme Production from T. longibrachiatum LMBC 172: Culture with Tamarind Seeds in Agitated and Static Conditions
3.2.5. Scaling for the Bioreactor to Increase Enzyme Production
3.2.6. Sugar Profile
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Damasio, A.R.L.; Rubio, M.V.; Gonçalves, T.A.; Persinoti, G.F.; Segato, F.; Prade, R.A.; Contesini, F.J.; Souza, A.P.; Buckeridge, M.S.; Squina, F.M. Xyloglucan breakdown by endo-xyloglucanase family 74 from Aspergillus fumigatus. Appl. Microbiol. Biotechnol. 2017, 101, 2893–2903. [Google Scholar] [CrossRef]
- Ribeiro, L.F.C.; De Lucas, R.C.; Vitcosque, G.; Ribeiro, L.F.; Ward, R.J.; Rubio, M.V.; Damasio, A.R.L.; Squina, F.M.; Gregory, R.C.; Walton, P.H.; et al. A novel thermostable xylanase GH10 from Malbranchea pulchella expressed in Aspergillus nidulans with potential applications in biotechnology. Biotechnol. Biofuels 2014, 7, 115. [Google Scholar] [CrossRef] [Green Version]
- Pasin, T.M.; Benassi, V.M.; Heinen, P.R.; Damasio, A.R.L.; Cereia, M.; Jorge, J.A.; Polizeli, M.L.T.M. Purification and functional properties of a novel glucoamylase activated by manganese and lead produced by Aspergillus japonicus. Int. J. Biol. Macromol. 2017, 102, 779–788. [Google Scholar] [CrossRef] [PubMed]
- Ichinose, H.; Araki, Y.; Michikawa, M.; Harazono, K.; Yaoi, K.; Karita, S.; Kaneko, S. Characterization of an endo-processive-type xyloglucanase having a β-1,4-glucan-binding module and a endo-type xyloglucanase from Streptomyces avermitilis. Appl. Microbiol. Biotechnol. 2012, 78, 7939–7945. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buckeridge, M.S. The evolution of the Glicomic Codes of extracellular matrices. BioSystems 2017, 164, 112–120. [Google Scholar] [CrossRef] [PubMed]
- Polizeli, M.L.T.M.; Rizzatti, A.C.S.; Monti, R.; Terenzi, H.F.; Jorge, J.A.; Amorim, D.S. Xylanases from fungi: Properties and industrial applications. Appl. Microbiol. Biotechnol. 2005, 67, 577–591. [Google Scholar] [CrossRef] [PubMed]
- Bai, S.; Dong, C.; Zhu, J.; Zhang, Y.; Dai, H. Identification of a xyloglucan-specific endo-(1-4)-beta-D-glucanase inhibitor protein from apple (Mallus × domestica Borkh.) as a potential defense gene against Botryosphaeria dothidea. Plant. Sci. 2015, 231, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Damasio, A.R.L.; Ribeiro, L.F.C.; Ribeiro, L.F.; Furtado, G.P.; Segato, F.; Almeida, F.B.R.; Crivellari, A.C.; Buckeridge, M.S.; Souza, T.A.C.B.; Murakami, M.T.; et al. Functional characterization and oligomerization of recombinant xyloglucan-specific endo-β-1,4-glucanase (GH12) from Aspergillus niveus. Biochim. Biophys. Acta 2012, 1874, 461–467. [Google Scholar] [CrossRef]
- Binoj, J.S.; Raj, E.; Daniel, B.S.S. Comprehensive characterization of industrially discarded fruit fiber, Tamarindus indica L. as a potential eco-friendly bioreinforcement for polymer composite. J. Clean. Prod. 2017, 142, 1321–1331. [Google Scholar] [CrossRef]
- Berezina, O.V.; Herlet, J.; Rykov, S.V.; Komberger, P.; Zavyalov, A.; Kozlov, D.; Sakhibgaraeva, L.; Krestyanova, I.; Schwarz, W.H.; Zverlov, V.V.; et al. Thermostable multifunctional GH74 xyloglucanase from Myceliophtora thermophila: High-level expression in Pichia pastoris and characterization of the recombinant protein. Appl. Microbiol. Biotechnol. 2017, 101, 5653–5666. [Google Scholar] [CrossRef]
- Tsuda, T.; Watanabe, M.; Ohshima, K.; Yamamoto, A.; Kawakishi, S.; Osawa, T. Antioxidative components isolated from the seed of tamarind (Tamarindus indica L.). J. Agric. Food Chem. 1994, 42, 2671–2674. [Google Scholar] [CrossRef]
- Lee, Y.; Langenheim, J.H. Systematics of the Genus hymenaea L. (Leguminosae, Caesalpinioideae, Deterieae); University of California Publications: Berkeley, CA, USA, 1975. [Google Scholar]
- Buckeridge, M.S.; Dietrich, S.M.C. Galactomannan from Brazilian legume seeds. Rev. Bras. Bot. 1990, 13, 109–112. [Google Scholar]
- Santos, H.P.; Buckeridge, M.S. The role of the storage carbon of cotyledons in the establishment of seedlings of Hymenaea courbaril under different light conditions. Ann. Bot. 2004, 94, 819–830. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alcântara, P.H.N.; Martim, L.; Silva, C.O.; Dietrich, S.M.C.; Buckeridge, M.S. Purification of a β-galactosidase from cotyledons of Hymenaea courbaril L. (Leguminosae). Enzyme properties and biological function. Plant. Physiol. Biochem. 2006, 44, 619–627. [Google Scholar] [CrossRef] [PubMed]
- Furtado, G.P.; Santos, C.R.; Cordeiro, R.L.; Ribeiro, L.F.; Moraes, L.A.B.; Damásio, A.R.L.; Polizeli, M.L.T.M.; Lourenzoni, M.R.; Murakami, M.T.; Ward, R.J. Enhanced xyloglucan-specific endo-β-1,4-glucanase efficiency in an engineered CBM-XEGA chimera. Appl. Microbiol. Biotechnol. 2015, 99, 5095–5107. [Google Scholar] [CrossRef] [PubMed]
- Ragauskas, A.J.; Beckham, G.T.; Biddy, M.J.; Chandra, R.; Chen, F.; Davis, M.F.; Davison, B.H.; Dixon, R.A.; Gilna, R.A.; Keller, M.; et al. Lignin valorization: Improving lignin processing in the biorefinery. Science 2014, 344, 1–12. [Google Scholar] [CrossRef]
- Li, Y.T.; Rouland, C.; Benedetti, M.; Li, F.; Lavelle, P.; Dai, J. Microbial biomass, enzyme and mineralization activity in relation to soil organic C, N and P turnover influenced by acid metal stress. Soil Biol. Biochem. 2009, 41, 969–977. [Google Scholar] [CrossRef]
- Rytioja, J.; Hildén, K.; Yuzon, J.; Hatakka, A.; de Vries, R.; Mäkelä, M.R. Plant-polysaccharide-degrading enzymes from Basidiomycetes. Microbiol. Mol. Biol. Rev. 2014, 78, 614–649. [Google Scholar] [CrossRef] [Green Version]
- Almeida, P.Z.D.; Pereira, M.G.; Carvalho, C.C.D.; Heinen, P.R.; Ziotti, L.S.; Messias, J.M.; Jorge, J.A.; Polizeli, M.L.T.M. Bioprospection and characterization of the amylolytic activity by filamentous fungi from Brazilian Atlantic Forest. Biota Neotrop 2017, 17. [Google Scholar] [CrossRef] [Green Version]
- Guimarães, L.H.S.; Peixoto-Nogueira, S.C.; Michelin, M.; Rizzatti, A.C.S.; Sandrim, V.C.; Zanoelo, F.F.; Aquino, A.C.M.M.; Barbosa Junior, A.; Polizeli, M.L.T.M. Screening of filamentous fungi for production of enzymes of biotechnological interest. Braz. J. Microbiol. 2006, 37, 474–480. [Google Scholar] [CrossRef]
- Oliveira, T.B.; Lucas, R.C.; Scarcella, A.S.A.; Contato, A.G.; Pasin, T.M.; Martinez, C.A.; Polizeli, M.L.T.M. Fungal communities differentially respond to warming and drought in tropical grassland soil. Mol. Ecol. 2020, 29, 1550–1559. [Google Scholar] [CrossRef] [PubMed]
- Barrett, C.; Lybbert, T. Is Bioprospecting a Viable Strategy for Conserving Tropical Ecosystems? In Environmental & Resource Economics; Cornell University: Ithaca, NY, USA, 1999. [Google Scholar]
- Emerson, R. An experimental study of the life cycles and taxonomy of Allomyces. Lloydia 1941, 4, 77–144. [Google Scholar]
- Contato, A.G.; Brugnari, T.; Sibin, A.P.A.; Buzzo, A.J.R.; Sá-Nakanishi, A.B.; Bracht, L.; Bersani-Amado, C.A.; Peralta, R.M.; Souza, C.G.M. Biochemical properties and effects on mitochondrial respiration of aqueous extracts of Basidiomycete mushrooms. Cell Biochem. Biophys. 2020, 78, 111–119. [Google Scholar] [CrossRef] [PubMed]
- White, T.J. PCR Protocols: A guide to method and applications. In Amplification and Direct Sequencing of Fungal Ribosomal rRNA Genes for Phylogenetics; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: San Diego, CA, USA, 1990; pp. 315–352. [Google Scholar]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/N.T. Nucleic Acids Res. 1999, 41, 95–98. [Google Scholar]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [Green Version]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef] [PubMed]
- Kimura, M. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khanna, P.; Sundari, S.S.; Kumar, N.J. Production, isolation and partial purification of xylanases from an Aspergillus sp. World J. Microbiol. Biotechnol. 1995, 11, 242–243. [Google Scholar] [CrossRef]
- Miller, G.L. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal. Chem. 1959, 31, 426–428. [Google Scholar] [CrossRef]
- Biely, P.; Puls, J.; Schneider, H. Acetyl xylan esterases in fungal cellulolytic systems. Febs. Lett. 1985, 186, 80–84. [Google Scholar] [CrossRef] [Green Version]
- Kersters-Hilderson, H.; Claeyssens, M.; Van Doorlaer, E.; Saman, E.; De Bruyne, C.K. beta-D-xylosidase from Bacillus pumilus. Methods Enzymol. 1982, 83, 631–639. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Michelin, M.; Teixeira, J.A. Liquid hot water pretreatment of multi feedstocks and enzymatic hydrolysis of solids obtained thereof. Bioresour. Technol. 2016, 216, 862–869. [Google Scholar] [CrossRef] [Green Version]
- Lima, M.S.; Damasio, A.R.L.; Crnkovic, P.M.; Pinto, M.R.; Silva, A.M.; Silva, J.C.; Segato, F.; Lucas, R.C.; Jorge, J.A.; Polizeli, M.L.T.M. Co-cultivation of Aspergillus nidulans recombinant strains produce an enzymatic cocktail as an alternative to alkaline sugarcane bagasse pretreatment. Front. Microbiol. 2016, 7, 583. [Google Scholar] [CrossRef] [PubMed]
- Henriques, O.K. Characterization of the Natural Vegetation in Ribeirão Preto, SP: Bases for Conservation. Ph.D. Thesis, Universidade de São Paulo (USP), São Paulo, Brazil, 2003. [Google Scholar]
- Bastos, R.C. Tecnologia das Fermentações: Fundamentos de Bioprocessos. Ph.D. Thesis, Collection UAB-UFScar, São Carlos, Brazil, 2010. [Google Scholar]
- Pereira, S.S. Compostos Bioativos e Caracterização de Sementes de Frutas Tropicais. Master’s Thesis, Universidade Federal de Lavras, Lavras, Brazil, 2017. [Google Scholar]
- Mori, F.A.; Mendes, L.M.; Trugilho, P.F. Utilização de eucaliptos e de madeiras nativas no armazenamento da aguardente de cana-de-açúcar. Food Sci. Technol. 2003, 23, 396–400. [Google Scholar] [CrossRef] [Green Version]
- Scarcella, A.S.A.; Pasin, T.M.; Lucas, R.C.; Ferreira-Nozawa, M.S.; Oliveira, T.B.; Contato, A.G.; Grandis, A.; Buckeridge, M.S.; Polizeli, M.L.T.M. Holocellulase production by filamentous fungi: Potential in the hydrolysis of energy cane and other sugarcane varities. Biomass Convers Biorefin. 2021. [Google Scholar] [CrossRef]



| Enzyme | Under Agitation | Static | ||||||
|---|---|---|---|---|---|---|---|---|
| 24 h | 48 h | 72 h | 96 h | 24 h | 48 h | 72 h | 96 h | |
| cellobiohydrolase | 4.6 ± 0 | 19.7 ± 1.2 | 25.0 ± 1.0 | 33.0 ± 2.0 | 2.1 ± 0 | 10.7 ± 0.6 | 17.3 ± 2.5 | 7.7 ± 0.9 |
| endoglucanase | 8.0 ± 1.7 | 107.3 ± 4.6 | 65.0 ± 9.5 | 53.0 ± 1.7 | 17.0 ± 2.0 | 77.3 ± 3.8 | 78.7 ± 0.6 | 84.3 ± 3.5 |
| β-D-glucosidase | 6.0 ± 0.3 | 51.0 ± 0 | 73.0 ± 0.6 | 69.0 ± 0.9 | 5.7 ± 0.3 | 11.0 ± 1.7 | 24.3 ± 2.9 | 25.0 ± 1.0 |
| β-glucanase | 32.0 ± 4.6 | 159.3 ± 6.7 | 93.6 ± 3.1 | 75.3 ± 11.9 | 5.7 ± 0.9 | 57.0 ± 9.5 | 109.0 ± 7.9 | 71.0 ± 11.1 |
| lichenase | 70.3 ± 9.5 | 299.0 ± 8.2 | 192.7 ± 9.5 | 90.3 ± 11.0 | 157.0 ± 0.9 | 245.0 ± 24.6 | 159.7 ± 3.5 | 145.3 ± 10.1 |
| endo-1,4-β-xylanase | 218.7 ± 44.5 | 893.7 ± 69.4 | 1043.3 ± 28.0 | 886.3 ± 20.6 | 31.0 ± 1.0 | 722.0 ± 12.5 | 845.3 ± 3.7 | 936.7 ± 1.2 |
| β-D-xylosidase | 3.0 ± 0.3 | 3.7 ± 0.4 | 4.8 ± 0 | 4.9 ± 0.5 | 3.1 ± 0.4 | 2.7 ± 0 | 2.0 ± 0.4 | 1.9 ± 0.4 |
| α-L-arabinofuranosidase | 3.5 ± 0.2 | 4.9 ± 1.1 | 7.0 ± 0.8 | nd | 1.5 ± 0.5 | 2.9 ± 0.2 | 5.9 ± 1.1 | 9.0 ± 1.2 |
| acetyl xylan esterase | nd | 19.0 ± 2.0 | 15.0 ± 1.0 | 5.3 ± 1.9 | nd | 1.6 ± 0.9 | 15.7 ± 1.2 | 4.2 ± 1.6 |
| endo-1,5-α-L-arabinanase | 81.0 ± 6.2 | 130.3 ± 16.1 | 105.0 ± 1.0 | 95.3 ± 2.5 | 55.5 ± 3.5 | 180.0 ± 6.2 | 158.0 ± 4.6 | 71.3 ± 4.0 |
| β-D-galactosidase | nd | 6.1 ± 0 | 11.3 ± 0.6 | 9.6 ± 1.2 | 2.7 ± 0.3 | 10.3 ± 0.6 | 15.0 ± 0 | 24.0 ± 1.0 |
| xyloglucanase | 22.3 ± 2.6 | 180.7 ± 18.2 | 210.0 ± 13.1 | 138.0 ± 18.0 | nd | 177.7 ± 3.5 | 311.0 ± 9.5 | 274.3 ± 5.1 |
| mannanase | 179.7 ± 30.0 | 265.0 ± 25.9 | 163.3 ± 7.2 | 112.7 ± 4.0 | 184.3 ± 1.2 | 247.3 ± 6.4 | 236.0 ± 2.6 | 226.7 ± 8.7 |
| polygalacturonase | 117.7 ± 0.6 | 133.3 ± 7.2 | 213.7 ± 4.0 | 56.7 ± 7.8 | 24.3 ± 4.2 | 138.7 ± 14.3 | 100.0 ± 6.0 | 88.7 ± 9.7 |
| Enzyme | Under Agitation | Static | ||||||
|---|---|---|---|---|---|---|---|---|
| 24 h | 48 h | 72 h | 96 h | 24 h | 48 h | 72 h | 96 h | |
| cellobiohydrolase | 2.3 ± 0.5 | 4.0 ± 0.6 | 5.0 ± 0.3 | 3.8 ± 0.7 | 1.4 ± 0 | 3.1 ± 0.3 | 3.4 ± 0.2 | nd |
| endoglucanase | 23.0 ± 10.6 | 39.3 ± 6.7 | 36.7 ± 5.0 | 23.0 ± 10.1 | 27.0 ± 4.4 | 43.7 ± 8.7 | 38.3 ± 1.2 | 27.7 ± 4.2 |
| β-D-glucosidase | 0.9 ± 0.5 | 11.7 ± 0.6 | 13.3 ± 1.5 | 16.7 ± 1.2 | 7.3 ± 0.7 | 19.0 ± 1.0 | 26.7 ± 0.6 | 19.3 ± 1.2 |
| β-glucanase | 21.0 ± 1.5 | 103.0 ± 8.2 | 136.7 ± 6.1 | 118.3 ± 6.1 | 25.7 ± 3.2 | 41.3 ± 5.7 | 49.3 ± 1.5 | 66.3 ± 7.6 |
| lichenase | nd | 7.0 ± 3.1 | 86.3 ± 12.0 | nd | 73.7 ± 19.3 | 113.0 ± 6.9 | 101.7 ± 21.7 | 58.7 ± 2.5 |
| endo-1,4-β-xylanase | 372.0 ± 12.1 | 649.3 ± 9.7 | 862.0 ± 10.8 | 660.0 ± 33.4 | 127.3 ± 3.9 | 363.7 ± 5.9 | 550.0 ± 8.9 | 701.7 ± 62.9 |
| β-D-xylosidase | 1.6 ± 0.5 | 3.1 ± 0.5 | 3.1 ± 1.0 | 1.4 ± 0.2 | 1.2 ± 0.4 | 2.1 ± 0.5 | 3.9 ± 1.3 | 4.3 ± 0.7 |
| α-L-arabinofuranosidase | 0.9 ± 0.1 | 5.3 ± 0.3 | 4.4 ± 0.2 | 3.6 ± 0.3 | 1.7 ± 0.8 | 2.3 ± 0.6 | 1.5 ± 0.9 | 4.4 ± 0.7 |
| acetyl xylan esterase | nd | 13.3 ± 2.3 | 19.0 ± 1.0 | 47.0 ± 0.7 | nd | 2.6 ± 0.9 | 39.0 ± 3.0 | 43.3 ± 0.6 |
| endo-1,5-α-L-arabinanase | 27.3 ± 10.0 | 51.7 ± 13.4 | 86.0 ± 13.2 | 23.3 ± 9.6 | 30.3 ± 7.5 | 34.3 ± 7.6 | 34.7 ± 9.9 | 9.3 ± 0.7 |
| β-D-galactosidase | 2.5 ± 0.4 | 3.0 ± 0.1 | 2.6 ± 0.4 | 1.8 ± 0.4 | 0.5 ± 0.2 | 1.0 ± 0.2 | 1.0 ± 0.2 | 0.9 ± 0.5 |
| xyloglucanase | 11.7 ± 7.6 | 46.3 ± 5.5 | 74.0 ± 4.6 | 84.0 ± 9.5 | 18.7 ± 2.1 | 39.3 ± 6.0 | 51.3 ± 0.6 | 27.3 ± 6.5 |
| mannanase | 77.0 ± 5.3 | 54.3 ± 4.7 | 52.3 ± 2.6 | 1.3 ± 0.3 | 28.7 ± 4.9 | 56.0 ± 7.1 | 29.7 ± 6.1 | 35.7 ± 4.0 |
| polygalacturonase | 81.7 ± 6.4 | 54.0 ± 5.0 | 22.0 ± 2.4 | 23.0 ± 4.2 | 26.7 ± 4.0 | 68.5 ± 6.4 | 85.7 ± 11.0 | 58.3 ± 1.5 |
| Enzyme | Under Agitation | Static | ||||||
|---|---|---|---|---|---|---|---|---|
| 24 h | 48 h | 72 h | 96 h | 24 h | 48 h | 72 h | 96 h | |
| cellobiohydrolase | 2.3 ± 0.2 | 3.8 ± 1.0 | 1.4 ± 0.7 | 0.5 ± 0.2 | 0.6 ± 0.3 | 1.4 ± 0.4 | 2.3 ± 0.5 | 1.1 ± 0.1 |
| endoglucanase | 11.0 ± 4.4 | 54.7 ± 4.7 | 45.7 ± 7.5 | 18.7 ± 6.5 | 24.7 ± 1.5 | 67.3 ± 0.6 | 50.7 ± 11.1 | 50.3 ± 9.8 |
| β-D-glucosidase | 9.3 ± 2.3 | 10.0 ± 0 | 11.0 ± 1.0 | 8.7 ± 1.2 | 3.4 ± 0.2 | 5.1 ± 0.2 | 6.3 ± 0.3 | 7.0 ± 0.6 |
| β-glucanase | 103.0 ± 1.5 | 177.0 ± 14.1 | 223.7 ± 19.1 | 59.3 ± 6.7 | 231.3 ± 20.5 | 161.7 ± 5.8 | 129.7 ± 10.5 | 41.3 ± 9.0 |
| lichenase | 70.3 ± 5.5 | 75.5 ± 10.5 | 97.3 ± 4.0 | 63.3 ± 17.0 | 131.3 ± 30.9 | 227.0 ± 5.8 | 244.0 ± 42.0 | 78.7 ± 17.0 |
| endo-1,4-β-xylanase | 30.0 ± 6.4 | 429.3 ± 9.5 | 492.3 ± 6.5 | 462.0 ± 4.4 | 31.0 ± 1.8 | 385.3 ± 33.0 | 772.3 ± 35.1 | 705.0 ± 23.8 |
| β-D-xylosidase | 1.2 ± 0 | 8.1 ± 0.9 | 10.0 ± 1.0 | 9.7 ± 1.2 | nd | 5.7 ± 0.2 | 19.0 ± 1.0 | 18.3 ± 2.1 |
| α-L-arabinofuranosidase | 1.2 ± 0.5 | 3.7 ± 0.3 | 4.9 ± 0.3 | 6.2 ± 0.5 | 1.6 ± 0.5 | 2.9 ± 0.2 | 8.7 ± 0.6 | 7.7 ± 0.6 |
| acetyl xylan esterase | 1.1 ± 0.1 | 7.1 ± 0.5 | 53.7 ± 2.9 | 8.6 ± 1.6 | nd | 6.1 ± 0.6 | nd | nd |
| endo-1,5-α-L-arabinanase | nd | 17.7 ± 5.1 | 32.7 ± 8.1 | nd | 3.3 ± 0.8 | 18.0 ± 7.9 | 1.8 ± 0.9 | nd |
| β-D-galactosidase | 2.3 ± 0.3 | 7.0 ± 0.7 | 9.3 ± 0.6 | 7.4 ± 0.7 | 2.8 ± 0.2 | 3.8 ± 0.4 | 9.0 ± 0 | 7.2 ± 1.8 |
| xyloglucanase | 11.7 ± 3.5 | 45.0 ± 7.2 | 61.7 ± 4.0 | 40.0 ± 5.3 | 14.1 ± 8.2 | 14.0 ± 7.2 | 69.7 ± 9.6 | 67.7 ± 12.3 |
| mannanase | 69.7 ± 3.1 | 99.7 ± 3.9 | 64.7 ± 10.5 | 2.7 ± 0.6 | 10.0 ± 3.6 | 71.0 ± 3.6 | 79.3 ± 5.9 | 102.7 ± 9.5 |
| polygalacturonase | 3.0 ± 0.8 | 4.0 ± 0.4 | 7.0 ± 1.2 | 8.1 ± 0.6 | 4.0 ± 0.9 | 7.3 ± 0.7 | 25.3 ± 18.5 | 10.5 ± 4.9 |
| Enzyme | Under Agitation | Static | ||||||
|---|---|---|---|---|---|---|---|---|
| 24 h | 48 h | 72 h | 96 h | 24 h | 48 h | 72 h | 96 h | |
| cellobiohydrolase | 3.5 ± 0.9 | 16.7 ± 1.2 | 1.8 ± 0.6 | 2.1 ± 0.7 | 0.3 ± 0.1 | 1.4 ± 0.4 | 2.5 ± 0.3 | 1.4 ± 1.1 |
| endoglucanase | 20.7 ± 3.2 | 33.0 ± 5.6 | 43.0 ± 5.1 | 50.7 ± 1.5 | nd | 44.0 ± 4.6 | 84.3 ± 10.7 | 61.7 ± 14.7 |
| β-D-glucosidase | 16.0 ± 0 | 42.7 ± 2.1 | 38.3 ± 1.5 | 23.7 ± 2.3 | 1.9 ± 0.3 | 8.2 ± 1.3 | 24.0 ± 2.6 | 16.3 ± 2.1 |
| β-glucanase | 181.7 ± 40.2 | 236.3 ± 19.4 | 389.0 ± 2.0 | 244.0 ± 6.0 | 22.7 ± 8.3 | 125.0 ± 9.5 | 185.7 ± 6.4 | 203.0 ± 11.1 |
| lichenase | 30.0 ± 10.4 | 60.3 ± 6.1 | 94.7 ± 32.9 | 84.0 ± 10.4 | 3.0 ± 0.9 | 36.0 ± 3.6 | 1.3 ± 0.3 | nd |
| endo-1,4-β-xylanase | 64.0 ± 9.8 | 225.3 ± 28.5 | 805.0 ± 32.7 | 725.0 ± 49.0 | 74.7 ± 2.5 | 776.0 ± 6.8 | 1011.0 ± 70.4 | 933.7 ± 26.7 |
| β-D-xylosidase | 2.4 ± 0.6 | 3.9 ± 0.1 | 12.3 ± 1.2 | 7.4 ± 0.7 | 0.9 ± 0.2 | 4.6 ± 0.2 | 5.0 ± 0.4 | 21.7 ± 3.8 |
| α-L-arabinofuranosidase | 1.5 ± 0.8 | 2.5 ± 0.1 | 4.1 ± 0.7 | nd | 0.3 ± 0.1 | 1.4 ± 0.8 | 2.6 ± 0.3 | 10.7 ± 1.2 |
| acetyl xylan esterase | 0.02 ± 0 | nd | 10.3 ± 1.5 | 7.2 ± 0.8 | 0.3 ± 0.1 | 10.0 ± 1.0 | 0.6 ± 0.2 | nd |
| endo-1,5-α-L-arabinanase | 27.3 ± 10.0 | 51.7 ± 13.4 | 86.0 ± 13.2 | 23.3 ± 9.6 | 27.0 ± 5.3 | 21.0 ± 83 | 6.0 ± 0.6 | nd |
| β-D-galactosidase | 0.8 ± 0.4 | 5.6 ± 0.4 | 6.7 ± 0.5 | 15.0 ± 0 | 0.8 ± 0 | 3.0 ± 0.3 | 5.9 ± 1.3 | 7.3 ± 0.4 |
| xyloglucanase | 13.0 ± 2.8 | 20.0 ± 5.7 | 40.0 ± 6.8 | 57.7 ± 4.3 | 12.3 ± 3.2 | 188.7 ± 9.0 | 325.7 ± 27.3 | 194.3 ± 9.0 |
| mannanase | 63.7 ± 6.1 | 100.0 ± 5.6 | 158.3 ± 18.9 | 35.0 ± 6.6 | 74.7 ± 4.0 | 98.0 ± 18.2 | 141.3 ± 18.9 | 39.3 ± 10.0 |
| polygalacturonase | 71.0 ± 6.0 | 246.0 ± 5.6 | 409.0 ± 18.0 | 60.0 ± 16.0 | 27.3 ± 2.8 | 352.0 ± 38.0 | 218.0 ± 9.2 | 190.3 ± 14.0 |
| Temperature Growth (°C) | Total Proteins Erlenmeyer Flasks * | Total Proteins Bioreactor * | |
|---|---|---|---|
| T. longibrachiatum | 30 | 18.2 ± 1.57 | 25.8 ± 1.91 |
| T. thermophilus | 50 | 21.3 ± 1.64 | 28.6 ± 2.03 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Contato, A.G.; de Oliveira, T.B.; Aranha, G.M.; de Freitas, E.N.; Vici, A.C.; Nogueira, K.M.V.; de Lucas, R.C.; Scarcella, A.S.d.A.; Buckeridge, M.S.; Silva, R.N.; et al. Prospection of Fungal Lignocellulolytic Enzymes Produced from Jatoba (Hymenaea courbaril) and Tamarind (Tamarindus indica) Seeds: Scaling for Bioreactor and Saccharification Profile of Sugarcane Bagasse. Microorganisms 2021, 9, 533. https://doi.org/10.3390/microorganisms9030533
Contato AG, de Oliveira TB, Aranha GM, de Freitas EN, Vici AC, Nogueira KMV, de Lucas RC, Scarcella ASdA, Buckeridge MS, Silva RN, et al. Prospection of Fungal Lignocellulolytic Enzymes Produced from Jatoba (Hymenaea courbaril) and Tamarind (Tamarindus indica) Seeds: Scaling for Bioreactor and Saccharification Profile of Sugarcane Bagasse. Microorganisms. 2021; 9(3):533. https://doi.org/10.3390/microorganisms9030533
Chicago/Turabian StyleContato, Alex Graça, Tássio Brito de Oliveira, Guilherme Mauro Aranha, Emanuelle Neiverth de Freitas, Ana Claudia Vici, Karoline Maria Vieira Nogueira, Rosymar Coutinho de Lucas, Ana Sílvia de Almeida Scarcella, Marcos Silveira Buckeridge, Roberto Nascimento Silva, and et al. 2021. "Prospection of Fungal Lignocellulolytic Enzymes Produced from Jatoba (Hymenaea courbaril) and Tamarind (Tamarindus indica) Seeds: Scaling for Bioreactor and Saccharification Profile of Sugarcane Bagasse" Microorganisms 9, no. 3: 533. https://doi.org/10.3390/microorganisms9030533
APA StyleContato, A. G., de Oliveira, T. B., Aranha, G. M., de Freitas, E. N., Vici, A. C., Nogueira, K. M. V., de Lucas, R. C., Scarcella, A. S. d. A., Buckeridge, M. S., Silva, R. N., & Polizeli, M. d. L. T. d. M. (2021). Prospection of Fungal Lignocellulolytic Enzymes Produced from Jatoba (Hymenaea courbaril) and Tamarind (Tamarindus indica) Seeds: Scaling for Bioreactor and Saccharification Profile of Sugarcane Bagasse. Microorganisms, 9(3), 533. https://doi.org/10.3390/microorganisms9030533