Diagnosis and Monitoring of Hepatitis B Virus Infection Using the Cobas® HBV Test for Use on the Cobas® 4800 System
Abstract
:1. Introduction
2. Materials and Methods
2.1. Standards
2.2. Clinical Specimens
2.3. HBV DNA Quantification
2.4. Statistical Analysis
3. Results
3.1. Specificity
3.2. Analytical Sensitivity
3.3. Precision
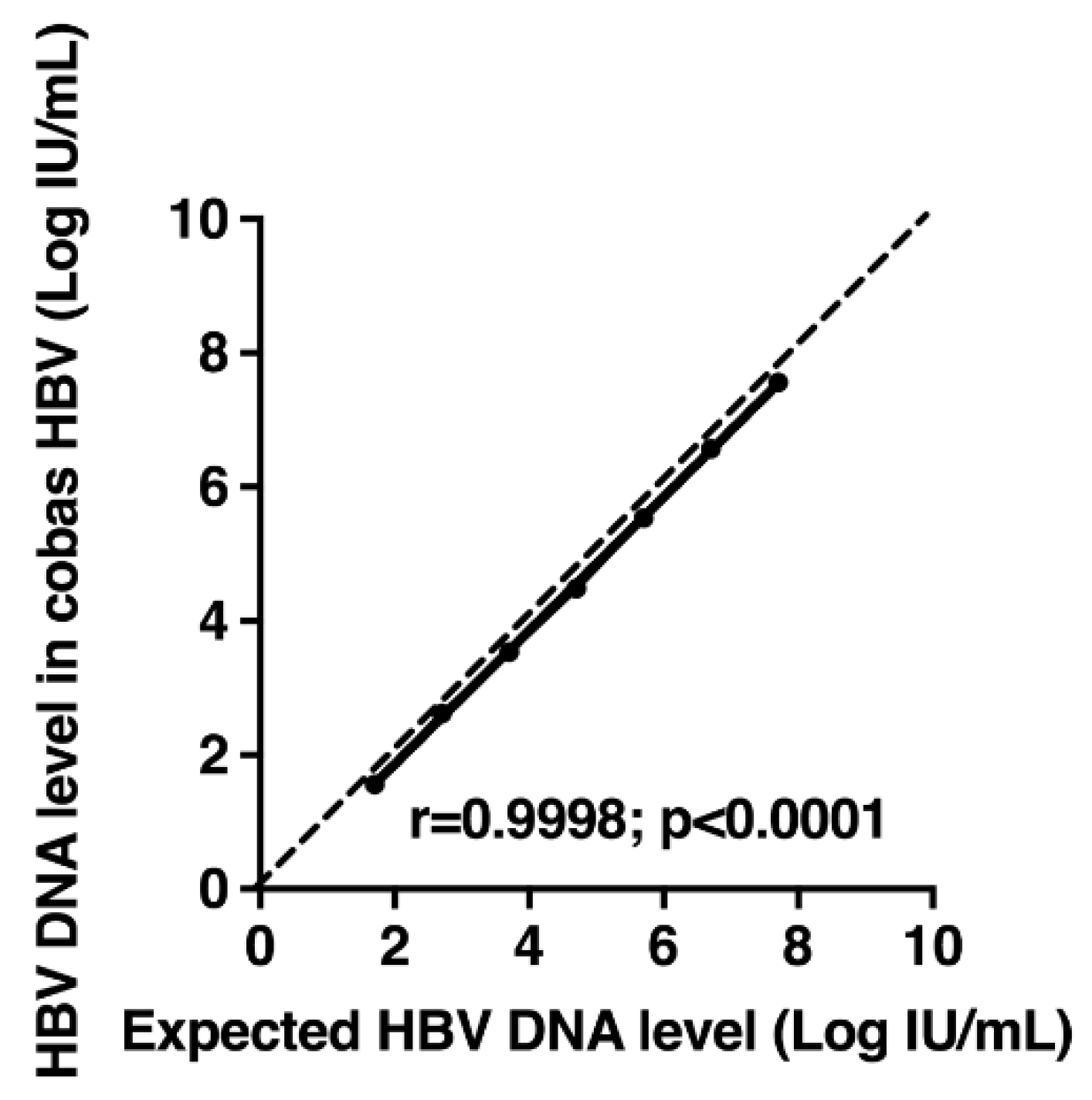
3.4. Linearity of a Standard Panel
3.5. Influence of Genotype on HBV DNA Quantification
3.6. Overall Percent Agreement for the Clinical Cutoffs of 2000 and 20,000 UI/mL
3.7. HBV DNA Monitoring in Patients Treated with Tenofovir-Containing Regimen
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Schweitzer, A.; Horn, J.; Mikolajczyk, R.T.; Krause, G.; Ott, J.J. Estimations of Worldwide Prevalence of Chronic Hepatitis B Virus Infection: A Systematic Review of Data Published Between 1965 and 2013. Lancet 2015, 386, 1546–1555. [Google Scholar] [CrossRef]
- Collaborators GBDC. The global, Regional, and National Burden of Cirrhosis by Cause in 195 Countries and Territories, 1990–2017: A Systematic Analysis for the Global Burden of Disease Study 2017. Lancet Gastroenterol. Hepatol. 2020, 5, 245–266. [Google Scholar] [CrossRef] [Green Version]
- Fitzmaurice, C.; Abate, D.; Allen, C.; Barber, R.M.; Barregard, L.; Bhutta, Z.A.; Brenner, H.; Satpathy, M. Global Burden of Disease Cancer C, Global, Regional, and National Cancer Incidence, Mortality, Years of Life Lost, Years Lived With Disability, and Disability-Adjusted Life-Years for 29 Cancer Groups, 1990 to 2017: A Systematic Analysis for the Global Burden of Disease Study. JAMA Oncol. 2019, 5, 1749–1768. [Google Scholar] [PubMed] [Green Version]
- Chang, T.T.; Liaw, Y.F.; Wu, S.S.; Schiff, E.; Han, K.H.; Lai, C.L.; Kreter, B. Long-term Entecavir Therapy Results in the Reversal of Fibrosis/cirrhosis and Continued Histological Improvement in Patients with Chronic Hepatitis B. Hepatology 2010, 52, 886–893. [Google Scholar] [CrossRef] [PubMed]
- Marcellin, P.; Gane, E.; Buti, M.; Afdhal, N.; Sievert, W.; Jacobson, I.M.; Heathcote, E.J. Regression of Cirrhosis during Treatment with Tenofovir Disoproxil Fumarate for Chronic Hepatitis B: A 5-year Open-label Follow-up Study. Lancet 2013, 381, 468–475. [Google Scholar] [CrossRef]
- European Association for the Study of the Liver. 2017 Clinical Practice Guidelines on the management of hepatitis B virus infection. J. Hepatol. 2017, 67, 370–398. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Terrault, N.A.; Lok, A.S.F.; McMahon, B.J.; Chang, K.M.; Hwang, J.P.; Jonas, M.M.; Wong, J.B. Update on Prevention, Diagnosis, and Treatment of Chronic Hepatitis B: AASLD 2018 Hepatitis B Guidance. Hepatology 2018, 67, 1560–1599. [Google Scholar] [CrossRef] [PubMed]
- Chevaliez, S.; Bouvier-Alias, M.; Laperche, S.; Hezode, C.; Pawlotsky, J.M. Performance of Version 2.0 of the Cobas AmpliPrep/Cobas TaqMan Real-time PCR assay for Hepatitis B virus DNA Quantification. J. Clin. Microbiol. 2010, 48, 3641–3647. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thibault, V.; Pichoud, C.; Mullen, C.; Rhoads, J.; Smith, J.B.; Bitbol, A.; Zoulim, F. Characterization of a New Sensitive PCR Assay for Quantification of Viral DNA Isolated from Patients with Hepatitis B Virus Infections. J. Clin. Microbiol. 2007, 45, 3948–3953. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yeh, Y.C.; Liu, C.J.; Kuo, R.N.; Lai, C.L.; Shau, W.Y.; Chen, P.J.; Lai, M.S. Association of Adjuvant Antiviral Therapy with Risk of Cancer Progression and Deaths in Patients with Hepatitis-B-virus-related Hepatocellular Carcinoma Following Curative Treatment: A Nationwide Cohort Study. PLoS ONE 2014, 9, e102051. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bonanzinga, S.; Chevaliez, S.; Onelia, F.; Jackson, K.; Pacenti, M.; Goldstein, E.; Galán, J.C.; Martínez-García, L.; Gunson, R. Multicenter clinical evaluation of alinity m HBV assay performance. J. Clin. Virol. 2020, 129, 104514. [Google Scholar] [CrossRef]
- Armbruster, D.A.; Pry, T. Limit of Blank, Limit of Detection and Limit of Quantitation. Clin. Biochem. Rev. 2008, 29 (Suppl. 1), 49–52. [Google Scholar]
- Chevaliez, S.; Dubernet, F.; Dauvillier, C.; Hezode, C.; Pawlotsky, J.M. The New Aptima HCV Quant Dx Real-time TMA Assay Accurately Quantifies Hepatitis C Virus Genotype 1-6 RNA. J. Clin. Virol. 2017, 91, 5–11. [Google Scholar] [CrossRef] [PubMed]
- Poiteau, L.; Wlassow, M.; Hezode, C.; Pawlotsky, J.M.; Chevaliez, S. Evaluation of the Xpert HBV Viral Load for Hepatitis B Virus Molecular Testing. J. Clin. Virol. 2020, 129, 104481. [Google Scholar] [CrossRef] [PubMed]
- Marcuccili, F.; Chevaliez, S.; Muller, T.; Colagrossi, L.; Abbondanza, G.; Beyser, K.; Ciotti, M. Multicenter Evaluation of the Cepheid Xpert HBV Viral Load test. Diagnostics 2021, 11, 297. [Google Scholar] [CrossRef] [PubMed]
- Maasoumy, B.; Bremer, B.; Lehmann, P.; Marins, E.G.; Michel-Treil, V.; Simon, C.O.; Wedemeyer, H. Commutability and concordance of four hepatitis B virus DNA assays in an international multicenter study. Therap. Adv. Gastroenterol. 2017, 10, 609–618. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aretzweiler, G.; Leuchter, S.; Garcia-Alvarez, M.; Simon, C.; Marins, E.; Paxinos, E.; Canchola, J.; Delgado, R.; Frontzek, A. Analytical Performance of Four Molecular Platforms Used for HIV-1, HBV and HCV Viral Load Determinations. Expert Rev. Mol. Diagn. 2019, 19, 941–949. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Hur, M.; Bae, E.; Lee, K.A.; Lee, W.I. Performance Evaluation of Cobas HBV Real-time PCR Assay on Roche Cobas 4800 System in Comparison with COBAS AmpliPrep/COBAS TaqMan HBV Test. Clin. Chem. Lab. Med. 2018, 56, 1133–1139. [Google Scholar] [CrossRef] [PubMed]
- Shin, K.H.; Lee, H.J.; Chang, C.L.; Kim, H.H. Performance of the Cobas Hepatitis B Virus (HBV) Test Using the Cobas 4800 System and Comparison of HBV DNA Quantification Ability between the COBAS AmpliPrep/COBAS TaqMan HBV Test Version 2.0 and Cobas HBV Test. J. Clin. Virol. 2018, 101, 47–51. [Google Scholar] [CrossRef] [PubMed]



| CAP/CTM HBV Test v2.0 | ||||
|---|---|---|---|---|
| cobas® HBV Test | <2000 IU/mL | ≥2000 IU/mL | Total | |
| <2000 IU/mL | 55 | 6 | 61 | |
| ≥2000 IU/mL | 3 | 57 | 60 | |
| Total | 58 | 63 | 121 | |
| <20,000 IU/mL | ≥20,000 IU/mL | Total | ||
| <20,000 IU/mL | 84 | 0 | 84 | |
| ≥20,000 IU/mL | 1 | 36 | 37 | |
| Total | 85 | 36 | 121 | |
| Alinity™ m HBV | ||||
|---|---|---|---|---|
| cobas® HBV Test | <2000 IU/mL | ≥2000 IU/mL | Total | |
| <2000 IU/mL | 39 | 5 | 44 | |
| ≥2000 IU/mL | 1 | 38 | 39 | |
| Total | 40 | 43 | 83 | |
| <20,000 IU/mL | ≥20,000 IU/mL | Total | ||
| <20,000 IU/mL | 62 | 0 | 62 | |
| ≥20,000 IU/mL | 0 | 21 | 21 | |
| Total | 62 | 21 | 83 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ortonne, V.; Wlassow, M.; Bouvier-Alias, M.; Melica, G.; Poveda, J.-D.; Laperche, S.; Pawlotsky, J.-M.; Chevaliez, S. Diagnosis and Monitoring of Hepatitis B Virus Infection Using the Cobas® HBV Test for Use on the Cobas® 4800 System. Microorganisms 2021, 9, 573. https://doi.org/10.3390/microorganisms9030573
Ortonne V, Wlassow M, Bouvier-Alias M, Melica G, Poveda J-D, Laperche S, Pawlotsky J-M, Chevaliez S. Diagnosis and Monitoring of Hepatitis B Virus Infection Using the Cobas® HBV Test for Use on the Cobas® 4800 System. Microorganisms. 2021; 9(3):573. https://doi.org/10.3390/microorganisms9030573
Chicago/Turabian StyleOrtonne, Valérie, Mélanie Wlassow, Magali Bouvier-Alias, Giovana Melica, Jean-Dominique Poveda, Syria Laperche, Jean-Michel Pawlotsky, and Stephane Chevaliez. 2021. "Diagnosis and Monitoring of Hepatitis B Virus Infection Using the Cobas® HBV Test for Use on the Cobas® 4800 System" Microorganisms 9, no. 3: 573. https://doi.org/10.3390/microorganisms9030573
APA StyleOrtonne, V., Wlassow, M., Bouvier-Alias, M., Melica, G., Poveda, J.-D., Laperche, S., Pawlotsky, J.-M., & Chevaliez, S. (2021). Diagnosis and Monitoring of Hepatitis B Virus Infection Using the Cobas® HBV Test for Use on the Cobas® 4800 System. Microorganisms, 9(3), 573. https://doi.org/10.3390/microorganisms9030573







