Plant Volatiles of Lettuce and Chicory Cultivated in Aquaponics Are Associated to Their Microbial Community
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection and Preparation
2.2. Plants Cultivation
2.3. Microbial Quantification by Culture-Dependent Method
2.4. Solid-Phase Microextraction–Gas Chromatography–Mass Spectrometry (SPME-GC-MS)
2.5. Statistical Analyses
3. Results and Discussion
3.1. General Aspects of the Volatilome Analysis through SPME-GC-MS
3.2. Volatilome Analysis of Lactuca Sativa
3.3. Volatilome Analysis of Cichorium Intybus
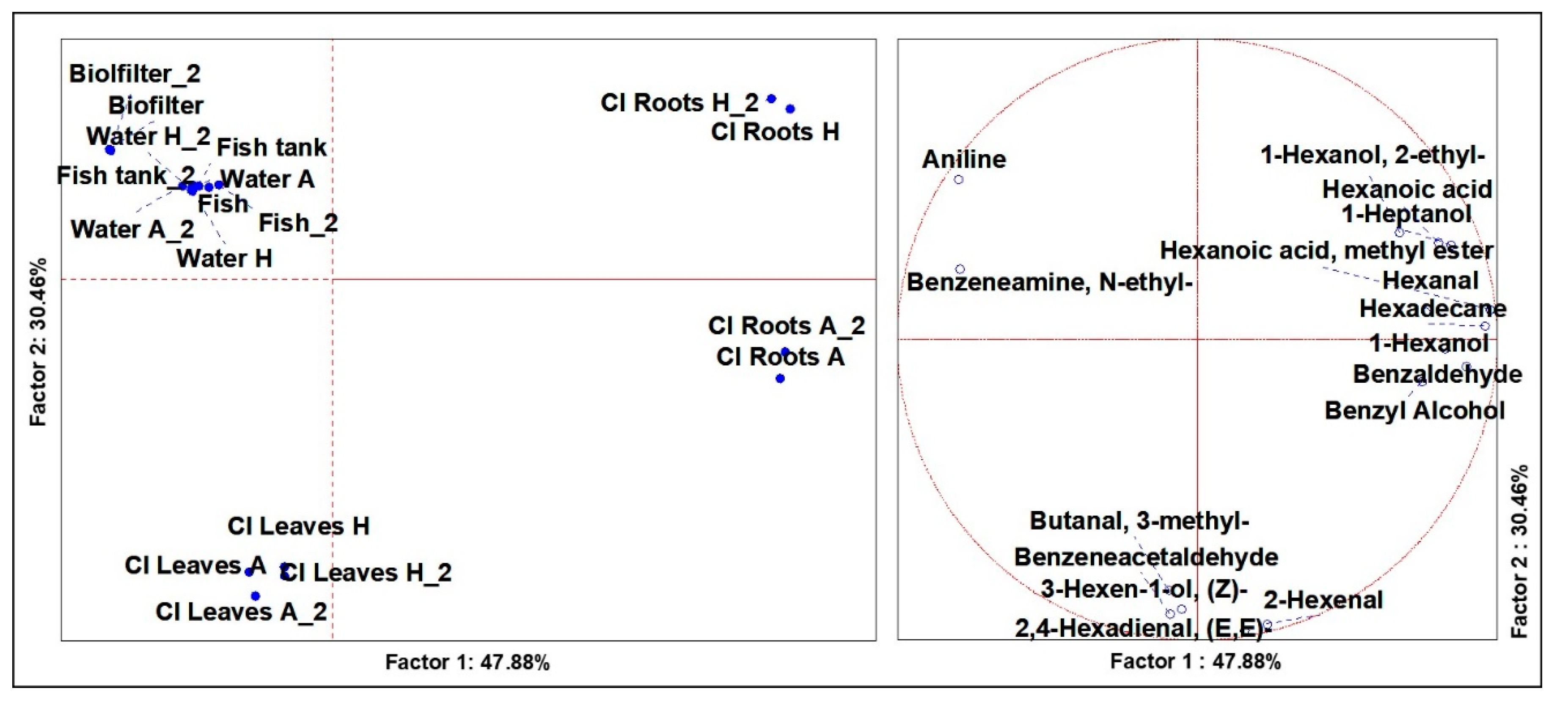
3.4. Volatilome Analysis of the System
3.5. Microbial Quantification
3.5.1. Microbial Quantification of the Phyllospheres
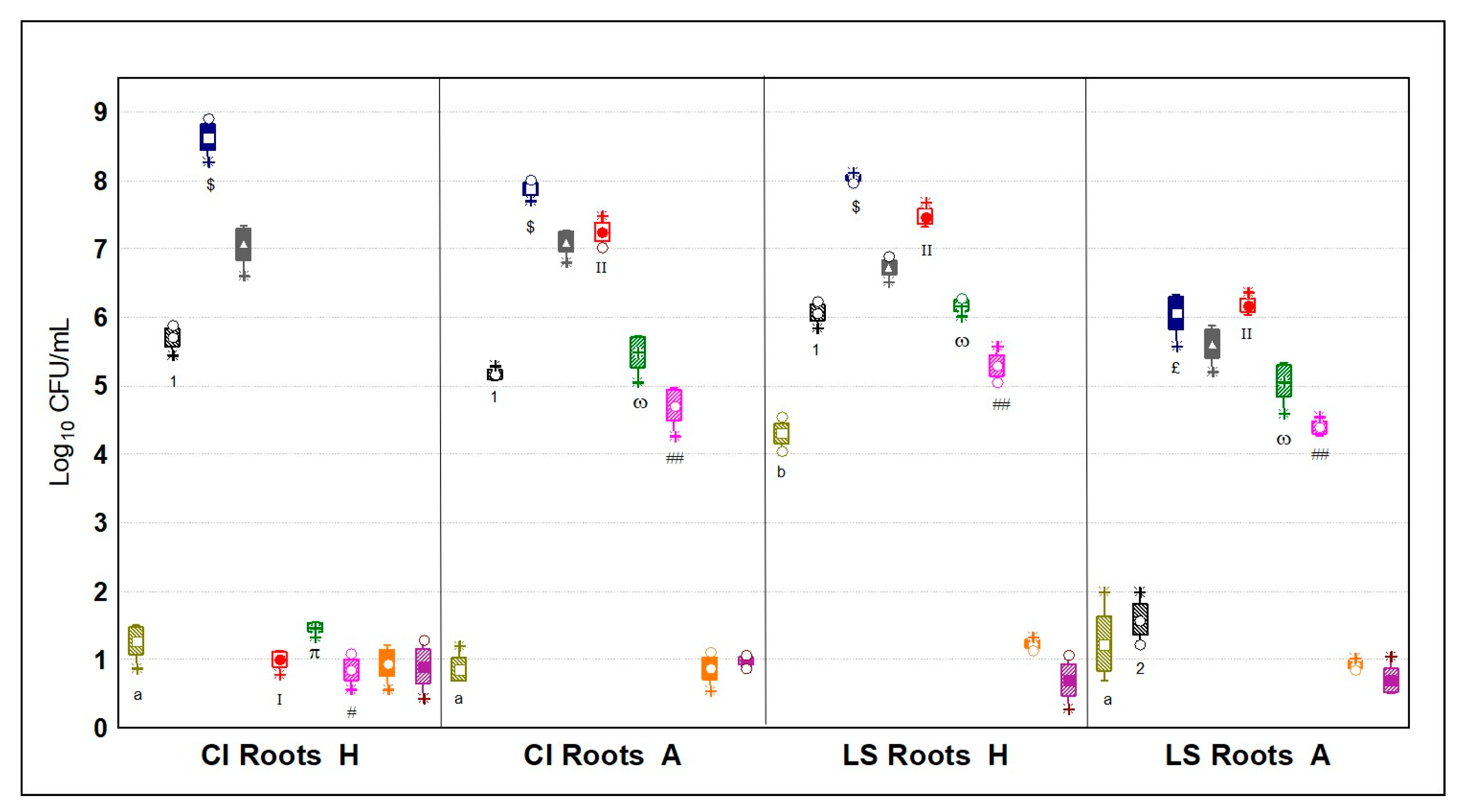
3.5.2. Microbial Quantification of the Rhizospheres
3.5.3. Microbial Quantification of the Cultivation Systems
3.6. Correlations among VOCs and Microbial Groups
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tilman, D.; Balzer, C.; Hill, J.; Befort, B.L. Global food demand and the sustainable intensification of agriculture. Proc. Natl. Acad. Sci. USA 2011, 108, 20260–20264. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Delaide, B.; Goddek, S.; Gott, J.; Soyeurt, H.; Jijakli, M.H. Lettuce (Lactuca sativa L. var. Sucrine) Growth Performance in Complemented Aquaponic Solution Outperforms Hydroponics. Water 2016, 8, 467. [Google Scholar] [CrossRef]
- Eck, M.; Sare, A.R.; Massart, S.; Schmautz, Z.; Junge, R.; Smits, T.H.M.; Jijakli, M.H. Exploring Bacterial Communities in Aquaponic Systems. Water 2019, 11, 260. [Google Scholar] [CrossRef] [Green Version]
- Fragozo, M.P.; Jacome, A.O.; Garcia, R.E.; Pacheco, T.I.; Hernandez, C.A.; Velazquez, O.R.V.; Trejo, G.J.F.; Gonzalez, G.R.G. Perspective for Aquaponic Systems: “Omic” Technologies for Microbial Community Analysis. BioMed Res. Int. 2015, 2015, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Schmautz, Z.; Graber, A.; Jaenicke, S.; Goesmann, A.; Junge, R.; Smits, T.H.M. Microbial diversity in different compartments of an aquaponics system. Arch. Microbiol. 2017, 199, 613–620. [Google Scholar] [CrossRef]
- Joyce, A.; Timmons, M.; Goddek, S.; Pentz, T. Bacterial Relationships in Aquaponics: New Research Directions. In Aquaponics Food Production Systems; Springer Science and Business Media LLC: New York, NY, USA, 2019; pp. 145–161. [Google Scholar]
- Bartelme, R.P.; Smith, M.C.; Villet, S.O.J.; Newton, R.J. Component Microenvironments and System Biogeography Structure Microorganism Distributions in Recirculating Aquaculture and Aquaponic Systems. mSphere 2019, 4, 19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Timmons, M.B.; Ebeling, J.M. Recirculating Aquaculture, 3rd ed.; Ithaca Publishing: New York, NY, USA, 2013; ISBN 978-0-9712646. [Google Scholar]
- Sahib, M.R.; Pervaiz, Z.H.; Williams, M.A.; Saleem, M.; DeBolt, S. Rhizobacterial species richness improves sorghum growth and soil nutrient synergism in a nutrient-poor greenhouse soil. Sci. Rep. 2020, 10, 1–13. [Google Scholar] [CrossRef]
- Yep, B.; Zheng, Y. Aquaponic trends and challenges–A review. J. Clean. Prod. 2019, 228, 1586–1599. [Google Scholar] [CrossRef]
- Kusano, M.; Redestig, H.; Hirai, T.; Oikawa, A.; Matsuda, F.; Fukushima, A.; Arita, M.; Watanabe, S.; Yano, M.; Tanase, H.K.; et al. Covering Chemical Diversity of Genetically-Modified Tomatoes Using Metabolomics for Objective Substantial Equivalence Assessment. PLoS ONE 2011, 6, 6989. [Google Scholar] [CrossRef] [Green Version]
- Zeng, W.; Hazebroek, J.; Beatty, M.; Hayes, K.; Ponte, C.; Maxwell, C.; Zhong, C.X. Analytical Method Evaluation and Discovery of Variation within Maize Varieties in the Context of Food Safety: Transcript Profiling and Metabolomics. J. Agric. Food Chem. 2014, 62, 2997–3009. [Google Scholar] [CrossRef]
- Sung, J.; Lee, S.; Lee, Y.; Ha, S.; Song, B.; Kim, T.; Waters, B.M.; Krishnan, H.B. Metabolomic profiling from leaves and roots of tomato (Solanum lycopersicum L.) plants grown under nitrogen, phosphorus or potassium-deficient condition. Plant Sci. 2015, 241, 55–64. [Google Scholar] [CrossRef]
- Garcia, C.J.; Villalba, G.R.; Garrido, Y.; Gil, M.I.; Barberán, T.F.A. Untargeted metabolomics approach using UPLC-ESI-QTOF-MS to explore the metabolome of fresh-cut iceberg lettuce. Metabolomics 2016, 12, 138. [Google Scholar] [CrossRef]
- Wang, Y.Q.; Hu, L.P.; Liu, G.M.; Zhang, D.S.; He, H.J. Evaluation of the Nutritional Quality of Chinese Kale (Brassica alboglabra Bailey) Using UHPLC-Quadrupole-Orbitrap MS/MS-Based Metabolomics. Molecules 2017, 22, 1262. [Google Scholar] [CrossRef] [Green Version]
- Nissen, L.; Di Carlo, E.; Gianotti, A. Prebiotic potential of hemp blended drinks fermented by probiotics. Food Res. Int. 2020, 131, 9029. [Google Scholar] [CrossRef] [PubMed]
- Saa, D.T.; Turroni, S.; Serrazanetti, D.I.; Rampelli, S.; Maccaferri, S.; Candela, M.; Severgnini, M.; Simonetti, E.; Brigidi, P.; Gianotti, A. Impact of Kamut® Khorasan on gut microbiota and metabolome in healthy volunteers. Food Res. Int. 2014, 63, 227–232. [Google Scholar] [CrossRef]
- Granato, D.; Calado, V.M.D.A.; Jarvis, B. Observations on the use of statistical methods in Food Science and Technology. Food Res. Int. 2014, 55, 137–149. [Google Scholar] [CrossRef]
- Nissen, L.; Rollini, M.; Picozzi, C.; Musatti, A.; Foschino, R.; Gianotti, A. Yeast-Free Doughs by Zymomonas mobilis: Evaluation of Technological and Fermentation Performances by Using a Metabolomic Approach. Microorganisms 2020, 8, 792. [Google Scholar] [CrossRef] [PubMed]
- Nissen, L.; Bordoni, A.; Gianotti, A. Shift of Volatile Organic Compounds (VOCs) in Gluten-Free Hemp-Enriched Sourdough Bread: A Metabolomic Approach. Nutrients 2020, 12, 1050. [Google Scholar] [CrossRef] [Green Version]
- Nissen, L.; Casciano, F.; Gianotti, A. Volatilome changes during probiotic fermentation of combined plant-based drinks. Food Funct. 2021. [Google Scholar] [CrossRef]
- Ioannidis, A.G.; Kerckhof, F.M.; Drif, Y.R.; Vanderroost, M.; Boon, N.; Ragaert, P.; De Meulenaer, B.; Devlieghere, F. Characterization of spoilage markers in modified atmosphere packaged iceberg lettuce. Int. J. Food Microbiol. 2018, 279, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Z.; Lu, J.R. Generation of Acetoin and Its Derivatives in Foods. J. Agric. Food Chem. 2014, 62, 6487–6497. [Google Scholar] [CrossRef] [PubMed]
- Hammami, R.; Fliss, I.; Corsetti, A. Editorial: Application of Protective Cultures and Bacteriocins for Food Biopreservation. Front. Microbiol. 2019, 10, 1561. [Google Scholar] [CrossRef] [Green Version]
- Tuersuntuoheti, T.; Wang, Z.; Zhang, M.; Asimi, S.; Liang, S.; Wang, Z.; Ren, X.; Sohail, A. Changes of microbial diversity and volatile compounds in edible and deteriorated Qingke barley fresh noodles stored at 25 °C. Int. J. Food Sci. Technol. 2020, 56, 885–896. [Google Scholar] [CrossRef]
- Fincheira, P.; Quiroz, A. Physiological response of Lactuca sativa exposed to 2-nonanone emitted by Bacillus sp. BCT Microbiol. Res. 2019, 219, 49–55. [Google Scholar] [CrossRef]
- Liu, B.; Wei, G.; Hu, Z.; Wang, G. Benzaldehyde Synthases Are Encoded by Cinnamoyl-CoA Reductase Genes in Cucumber (Cucumis sativus L.). bioRxiv 2019. [Google Scholar] [CrossRef] [Green Version]
- Lalas, S.; Athanasiadis, V.; Karageorgou, I.; Bozinou, E.; Dourtoglou, V.G. Study of the self-stabilization ability of Tzatziki (a traditional Greek ready-to-eat deli salad). Int. J. Food Stud. 2019, 8, 76–86. [Google Scholar] [CrossRef]
- Xu, J.; Liu, Y.; Zhu, C.; Jia, H.; Tian, C.; Ma, H.; Lv, G. NaCl Improves Suaeda salsa Aniline Tolerance in Wastewater. Sustainability 2020, 12, 7457. [Google Scholar] [CrossRef]
- Pothakos, V.; Snauwaert, C.; De Vos, P.; Huys, G.; Devlieghere, F. Psychrotrophic members of Leuconostoc gasicomitatum, Leuconostoc gelidum and Lactococcus piscium dominate at the end of shelf-life in packaged and chilled-stored food products in Belgium. Food Microbiol. 2014, 39, 61–67. [Google Scholar] [CrossRef] [PubMed]
- Schmautz, Z.; Loeu, F.; Liebisch, F.; Graber, A.; Mathis, A.; Griessler Bulc, T.; Junge, R. Tomato Productivity and Quality in Aquaponics: Comparison of Three Hydroponic Methods. Water 2016, 8, 533. [Google Scholar] [CrossRef] [Green Version]
- Bartelme, R.P.; Oyserman, B.O.; Blom, J.E.; Sepulveda-Villet, O.J.; Newton, R.J. Stripping Away the Soil: Plant Growth Promoting Microbiology Opportunities in Aquaponics. Front. Microbiol. 2018, 9, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Avis, T.J.; Gravel, V.; Antoun, H.; Tweddell, R.J. Multifaceted beneficial effects of rhizosphere microorganisms on plant health and productivity. Soil Biol. Biochem. 2008, 40, 1733–1740. [Google Scholar] [CrossRef]
- Birch, A.N.; Petersen, M.A.; Hansen, Å.S. The aroma profile of wheat bread crumb influenced by yeast concentration and fermentation temperature. LWT 2013, 50, 480–488. [Google Scholar] [CrossRef]
- Day, J.; Otwell, A.; Diener, C.; Tams, K.E.; Bebout, B.; Detweiler, A.M.; Lee, M.D.; Scott, M.T.; Ta, W.; Ha, M.; et al. Negative plant-microbiome feedback limits productivity in aquaponics. bioRxiv 2019. [Google Scholar] [CrossRef] [Green Version]
- Vandenberghe, L.P.D.S.; Garcia, L.M.B.; Rodrigues, C.; Camara, M.C.; Pereira, G.V.D.M.; De Oliveira, J.; Soccol, C.R. Potential applications of plant probiotic microorganisms in agriculture and forestry. AIMS Microbiol. 2017, 3, 629–648. [Google Scholar] [CrossRef]






Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nissen, L.; Casciano, F.; Gianotti, A. Plant Volatiles of Lettuce and Chicory Cultivated in Aquaponics Are Associated to Their Microbial Community. Microorganisms 2021, 9, 580. https://doi.org/10.3390/microorganisms9030580
Nissen L, Casciano F, Gianotti A. Plant Volatiles of Lettuce and Chicory Cultivated in Aquaponics Are Associated to Their Microbial Community. Microorganisms. 2021; 9(3):580. https://doi.org/10.3390/microorganisms9030580
Chicago/Turabian StyleNissen, Lorenzo, Flavia Casciano, and Andrea Gianotti. 2021. "Plant Volatiles of Lettuce and Chicory Cultivated in Aquaponics Are Associated to Their Microbial Community" Microorganisms 9, no. 3: 580. https://doi.org/10.3390/microorganisms9030580
APA StyleNissen, L., Casciano, F., & Gianotti, A. (2021). Plant Volatiles of Lettuce and Chicory Cultivated in Aquaponics Are Associated to Their Microbial Community. Microorganisms, 9(3), 580. https://doi.org/10.3390/microorganisms9030580






