Antioxidant and Anti-Inflammatory Properties of Recombinant Bifidobacterium bifidum BGN4 Expressing Antioxidant Enzymes
Abstract
1. Introduction
2. Materials and Methods
2.1. Optimum Conditions for Antioxidant Enzyme Activities
2.1.1. Culture of BGN4 and BGN4-SK
2.1.2. Preparation of Crude Enzyme Extract of BGN4-SK
2.1.3. Measurement of SOD and Catalase Activities
2.1.4. Optimization of Medium for SOD and Catalase Production by BGN4-SK
2.2. Preparation of Bacteria Culture and Cell Culture and Cell Viability Assay
2.2.1. Culture of BGN4 and BGN4-SK
2.2.2. Cell Lines and Culture Conditions
2.2.3. Cell Viability Assay
2.3. Antioxidant Activities of BGN4-SK in H2O2-Stimulated HT-29 Cells
2.3.1. Determination of Intracellular Radical Oxygen Species (ROS) Levels
2.3.2. Determination of Intracellular Antioxidant Enzyme Activity
2.4. Determination of Cytokine Levels in LPS-Stimulated HT-29 Cells and RAW 264.7 Cells
2.5. Statistical Analysis
3. Results and Discussion
3.1. Optimum Conditions for the Activities of SOD and Catalase
3.1.1. Effect of MnSO4 Concentrations on SOD Activity
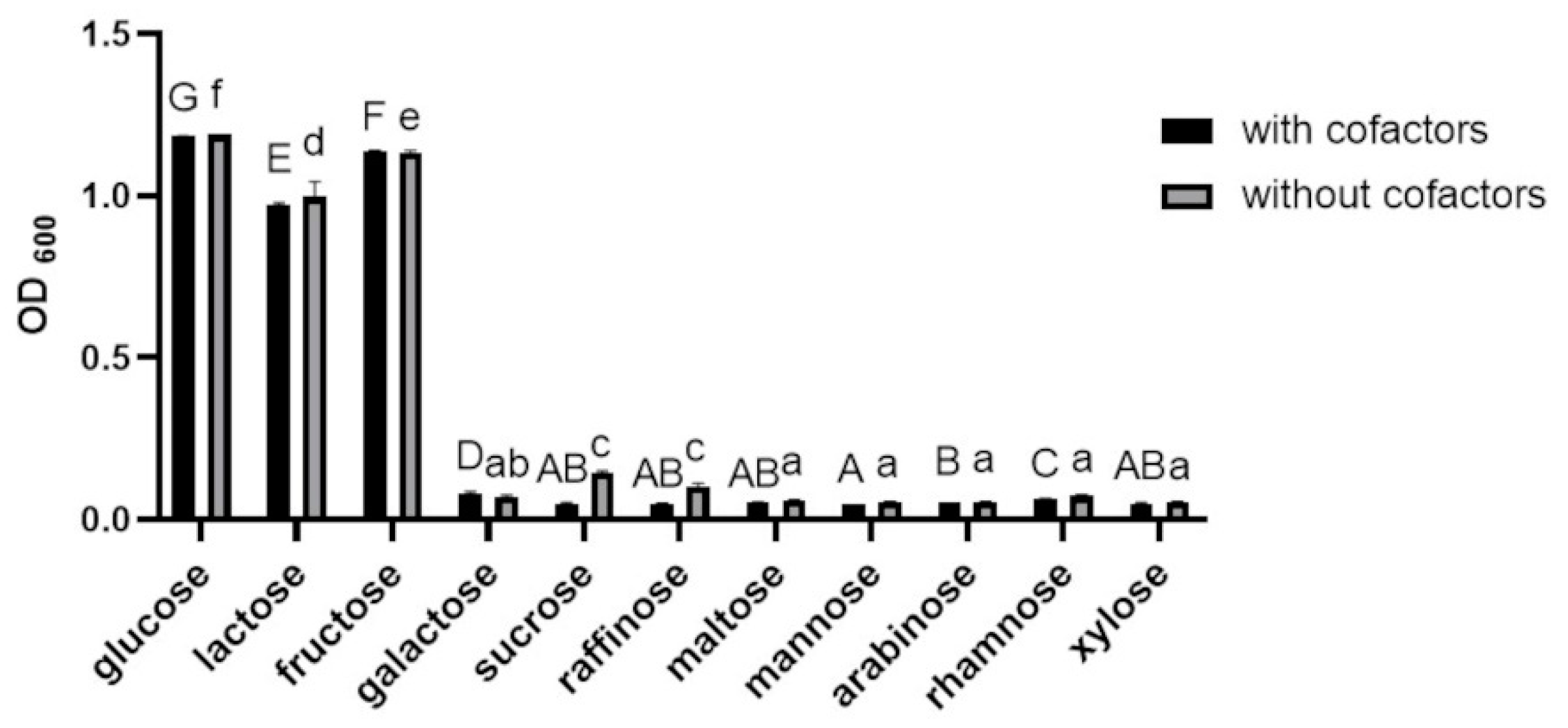
3.1.2. Effects of Various Carbon Sources and Cofactors on the Growth of BGN4-SK
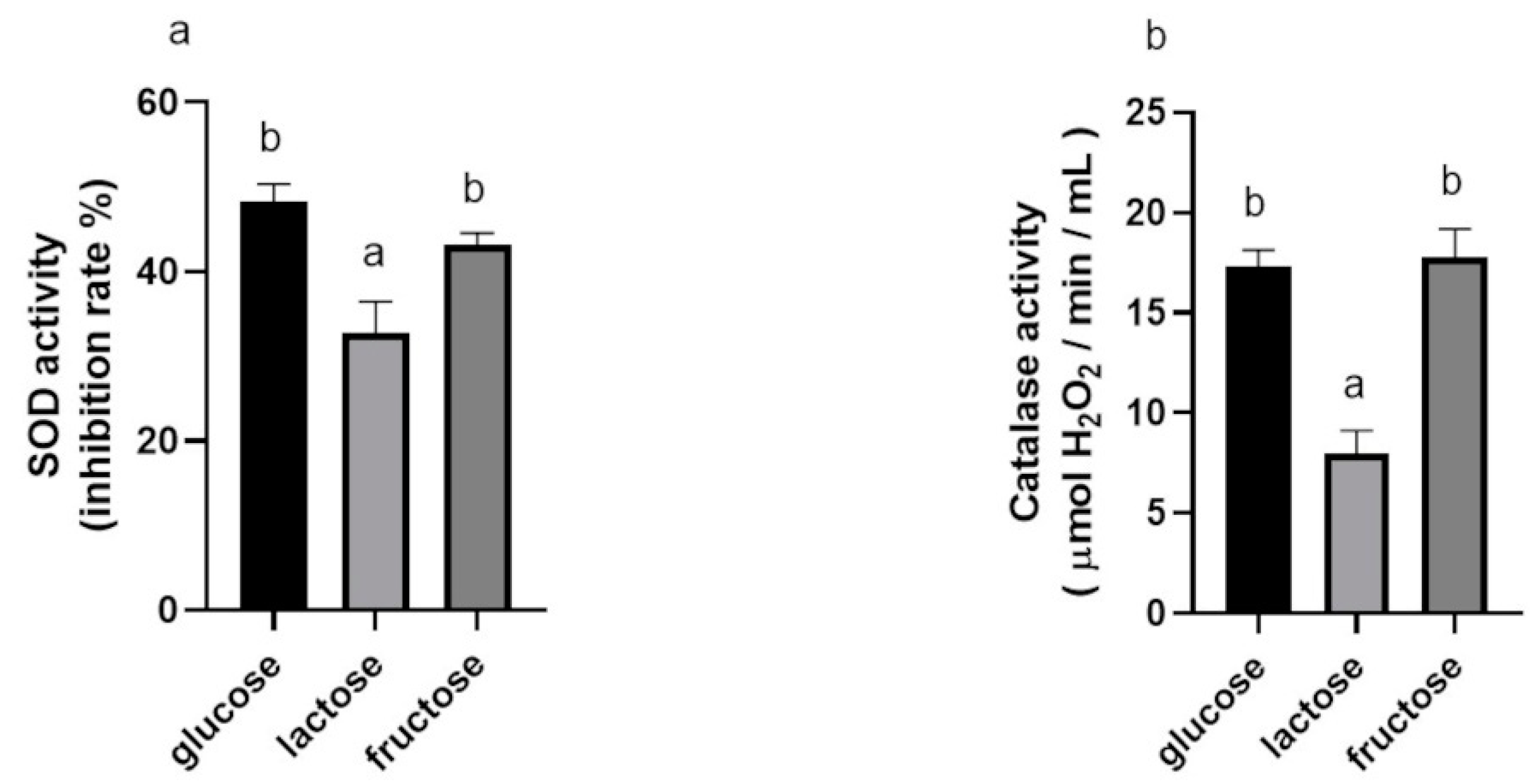
3.1.3. Effects of Carbon Sources on SOD and Catalase Activities of BGN4-SK
3.1.4. Effects of Incubation Times and Carbon Sources on SOD and Catalase Activities
3.2. Cell Viability
3.3. Antioxidant Activities of BGN4-SK in H2O2-Stimulated HT-29 Cells
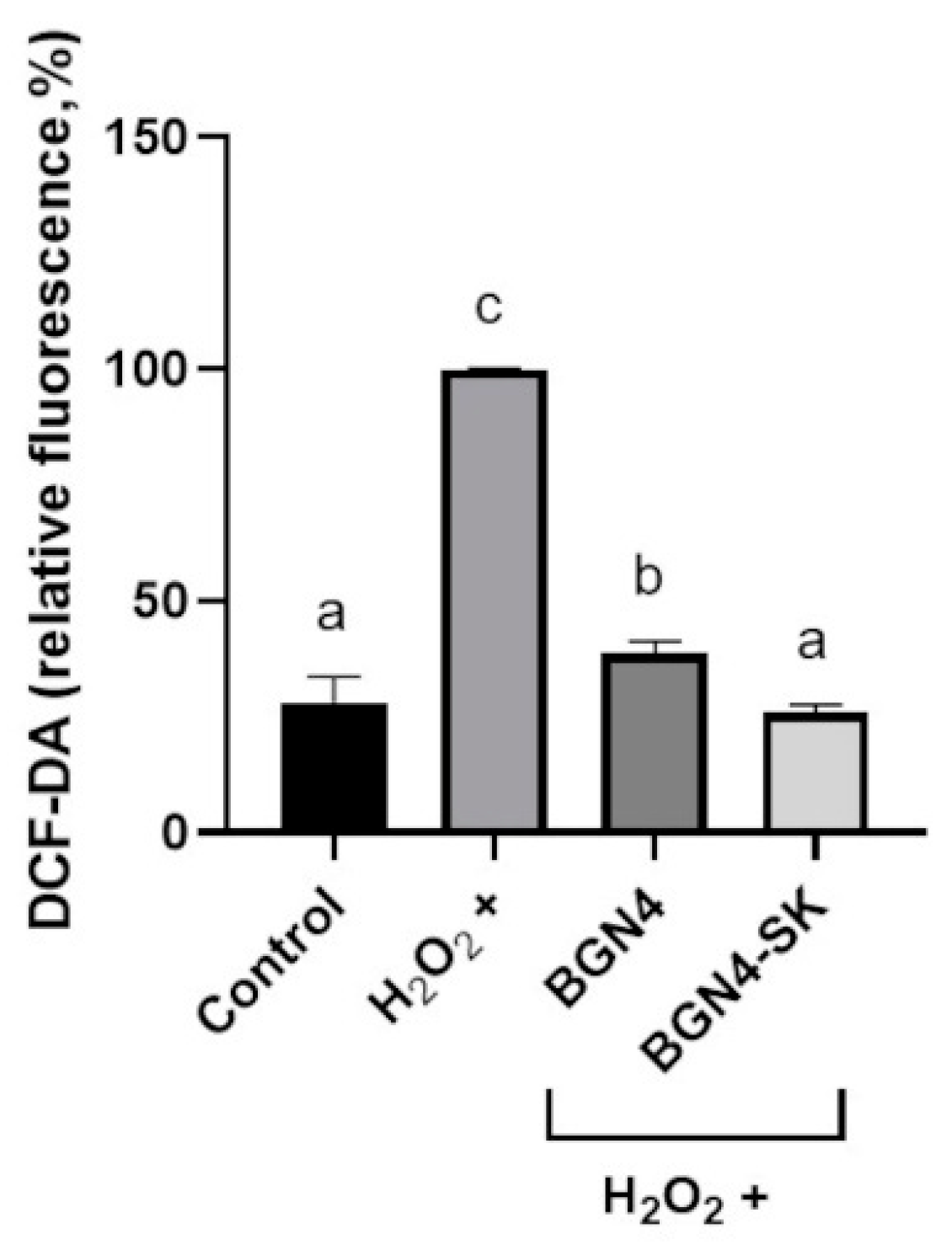
3.3.1. Intracellular ROS Levels
3.3.2. Intracellular Antioxidant Enzyme Activity
3.4. Anti-Inflammatory Activities of BGN4-SK in LPS-Stimulated RAW 264.7 Cells and HT-29 Cells
3.4.1. Inhibitory Effect of BGN4-SK on the Expression of the Pro-Inflammatory Cytokines IL-6 and TNF-α in LPS-Stimulated RAW 264.7 Cells
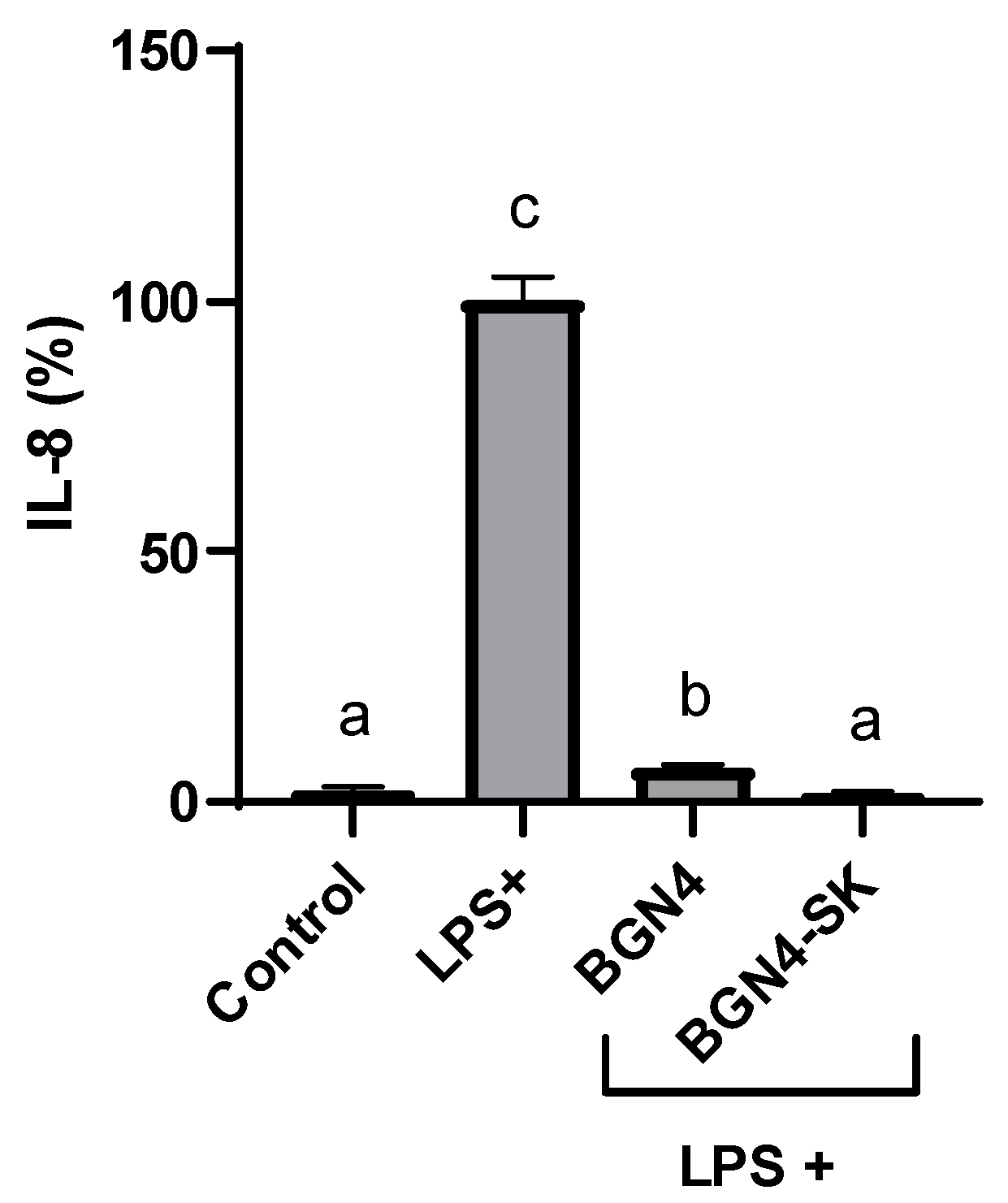
3.4.2. Inhibitory Effect of BGN4-SK on the Expression of IL-8 in LPS-Stimulated HT-29 Cells
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mishra, V.; Shah, C.; Mokashe, N.; Chavan, R.; Yadav, H.; Prajapati, J. Probiotics as potential antioxidants: A systematic review. J. Agric. Food. Chem. 2015, 63, 3615–3626. [Google Scholar] [CrossRef]
- Park, M. Construction of Recombinant Bifidobacterium bifidum BGN4 Expressing Bifidobacterial β-Galactosidase Gene and Application for Improvement of Lactose Intolerance. Ph.D. Thesis, Seoul National University, Seoul, Korea, 2019. [Google Scholar]
- LeBlanc, A.M.; Carmen, S.; Chatel, J.; Miyoshi, A.; Azevedo, V.; Langella, P.; Bermúdez-Humarán, L.G.; LeBlanc, G. Current review of genetically modified lactic acid bacteria for the prevention and treatment of colitis using murine models. Gastroenterol. Res. Pract. 2015, 2015, 1–8. [Google Scholar] [CrossRef]
- Carmen, S.; LeBlanc, A.; Martin, R.; Chain, F.; Langella, P.; Bermúdez-Humarán, L.; LeBlanc, J. Genetically engineered immunomodulatory Streptococcus thermophilus strains producing antioxidant enzymes exhibit enhanced antiinflammatory activities. Appl. Environ. Microbiol. 2014, 80, 869–877. [Google Scholar] [CrossRef] [PubMed]
- LeBlanc, J.; Carmen, S.; Miyoshi, A.; Azevedo, V.; Sesma, F.; Langella, P.; Bermúdez-Humarán, L.; Watterlot, L.; Perdigon, G.; LeBlanc, A.M. Use of superoxide dismutase and catalase producing lactic acid bacteria in TNBS induced Crohn’s disease in mice. J. Biotechnol. 2011, 151, 287–293. [Google Scholar] [CrossRef] [PubMed]
- Aoki, R.; Tsuchida, S.; Arai, Y.; Ohno, K.; Nishijima, T.; Mawatari, T.; Mikami, Y.; Ushida, K. Effect of Bifidobacterium animalis subsp. lactis GCL2505 on the physiological function of intestine in a rat model. Nutr. Food. Sci. 2016, 4, 782–790. [Google Scholar]
- Hong, N.; Ku, S.; Yuk, K.; Johnston, T.; Ji, G.; Park, M. Production of biologically active human interleukin-10 by Bifidobacterium bifidum BGN4. Microb. Cell Fact. 2021, 20, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Ku, S.; Park, M.; Ji, G.; You, H. Review on Bifidobacterium bifidum BGN4: Functionality and nutraceutical applications as a probiotic microorganism. IJMCFK Sci. 2016, 17, 1544. [Google Scholar] [CrossRef]
- Kim, M.; Ku, S.; Kim, S.Y.; Lee, H.H.; Jin, H.; Kang, S.; Li, R.; Jhnston, T.V.; Park, M.S.; Ji, G.E. Safety evaluations of Bifidobacterium bifidum BGN4 and Bifidobacterium longum BORI. Int. J. Mol. Sci. 2018, 19, 1422. [Google Scholar] [CrossRef] [PubMed]
- Zuo, F.; Yu, R.; Feng, X.; Khaskheli, G.; Chen, L.; Ma, H.; Chen, S. Combination of heterogeneous catalase and superoxide dismutase protects Bifidobacterium longum strain NCC2705 from oxidative stress. Appl. Microbiol. Biotechnol. 2014, 98, 7523–7534. [Google Scholar] [CrossRef]
- Zuo, F.; Chen, S.; Marcotte, H. Engineer probiotic bifidobacteria for food and biomedical applications—Current status and future prospective. Biotechnol. Adv. 2020, 45, 1–12. [Google Scholar] [CrossRef]
- Su, J.; Li, Z.; Liao, B.; Zhu, Y.; Zhang, X.; Wang, C.; He, J. Microcalorimetric study of the effect of manganese on the growth and metabolism in a heterogeneously expressing manganese-dependent superoxide dismutase (Mn-SOD) strain. J. Therm. Anal. Calorim. 2017, 130, 1407–1416. [Google Scholar] [CrossRef]
- Zhu, Y.; Wang, G.; Ni, H.; Xiao, A.; Cai, H. Cloning and characterization of a new manganese superoxide dismutase from deep-sea thermophile Geobacillus sp. EPT3. World J. Microbiol. Biotechnol. 2014, 30, 1347–1357. [Google Scholar] [CrossRef]
- Fee, J.A. Regulation of sod genes in Escherichia coli: Relevance to superoxide dismutase function. Mol. Microbiol. 1991, 5, 2599–2610. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Liu, G.; Chen, Y.; Huang, J.; Zhu, Y. Optimization of fermentation conditions in a shaker for the recombinant manganese superoxide dismutase from engineering E. coli. Jimei Univ. J. Nat. Sci. 2012, 17, 26–32. [Google Scholar]
- Angelis, M.; Gobbetti, M. Lactobacillus sanfranciscensis CB1: Manganese, oxygen, superoxide dismutase and metabolism. Appl. Microbiol. Biotechnol. 1998, 51, 358–363. [Google Scholar] [CrossRef]
- Albert, K.; Rani, A.; Sela, D.A. Comparative pangenomics of the mammalian gut commensal Bifidobacterium longum. Microorganisms 2019, 8, 7. [Google Scholar] [CrossRef]
- Park, M.; Park, M.S.; Ji, G.E. Cloning and heterologous expression of the β-galactosidase gene from Bifidobacterium longum RD47 in B. bifidum BGN4. J. Microbiol. Biotechnol. 2019, 29, 1717–1728. [Google Scholar] [CrossRef]
- Hwang, S. Utilization of Fucosylated Oligosaccharides by Bifidobacterium bifidum BGN4 and Expression of Cloned BGN4 α-L-Fucosidase Genes in E. coli. Master’s Thesis, Seoul National University, Seoul, Korea, 2015. [Google Scholar]
- Kim, Y.; Miller, C.D.; Anderson, A.J. Transcriptional regulation by iron of genes encoding iron- and manganese-superoxide dismutases from Pseudomonas putida. Gene 1999, 239, 129–135. [Google Scholar] [CrossRef]
- Deng, Y.; Xu, M.; Ji, D.; Agyei, D. Optimization of β-galactosidase production by batch cultures of Lactobacillus leichmannii 313. Fermentation 2019, 6, 1–17. [Google Scholar]
- Wang, Y.; Guo, Y.; Chen, H.; Wei, H.; Wan, C. Potential of Lactobacillus plantarum ZDY2013 and Bifidobacterium bifidum WBIN03 in relieving colitis by gut microbiota, immune, and anti-oxidative stress. NRC Res. Press 2018, 64, 327–337. [Google Scholar] [CrossRef] [PubMed]
- Riedel, C.; Foata, F.; Philippe, D.; Adolfsson, O.; Eikmanns, B.; Blum, S. Anti-inflammatory effects of bifidobacteria by inhibition of LPS-induced NF-κB activation. World J. Gastroenterol. 2006, 12, 3729–3735. [Google Scholar] [CrossRef] [PubMed]
- Baez, A.; Shiloach, J. Escherichia coli avoids high dissolved oxygen stress by activation of SoxRS and manganese superoxide dismutase. Microbial. Cell Fact. 2013, 12, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Mu, G.; Li, H.; Tuo, Y.; Zhang, Y. Antioxidative effect of Lactobacillus plantarum Y44 on 2,2′-azobis (2-methylpropionamidine) dihydrochloride (ABAP)-damaged Caco-2 cells. J. Dairy Sci. 2019, 102, 6863–6875. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Ji, G.; Ko, Y.; Jung, H.; Ustunol, Z.; Pestk, J. Potentiation of hydrogen peroxide, nitric oxide, and cytokine production in RAW 264.7 macrophage cells exposed to human and commercial isolates of Bifidobacterium. Int. J. Food Microbiol. 1999, 46, 231–241. [Google Scholar] [CrossRef]
- Srivastava, S.; Yadav, U.; Reddy, A.; Saxena, A.; Tammali, R.; Shoeb, M.; Ansari, N.; Bhatnagar, A.; Petrash, M.; Srivastava, S.; et al. Aldose reductase inhibition suppresses oxidative stress-induced inflammatory disorders. Chem. Biol. Interact. 2011, 191, 330–338. [Google Scholar] [CrossRef] [PubMed]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lin, Z.; Ku, S.; Lim, T.; Park, S.Y.; Park, M.S.; Ji, G.E.; O’Brien, K.; Hwang, K.T. Antioxidant and Anti-Inflammatory Properties of Recombinant Bifidobacterium bifidum BGN4 Expressing Antioxidant Enzymes. Microorganisms 2021, 9, 595. https://doi.org/10.3390/microorganisms9030595
Lin Z, Ku S, Lim T, Park SY, Park MS, Ji GE, O’Brien K, Hwang KT. Antioxidant and Anti-Inflammatory Properties of Recombinant Bifidobacterium bifidum BGN4 Expressing Antioxidant Enzymes. Microorganisms. 2021; 9(3):595. https://doi.org/10.3390/microorganisms9030595
Chicago/Turabian StyleLin, Zhaoyan, Seockmo Ku, Taehwan Lim, Sun Young Park, Myeong Soo Park, Geun Eog Ji, Keely O’Brien, and Keum Taek Hwang. 2021. "Antioxidant and Anti-Inflammatory Properties of Recombinant Bifidobacterium bifidum BGN4 Expressing Antioxidant Enzymes" Microorganisms 9, no. 3: 595. https://doi.org/10.3390/microorganisms9030595
APA StyleLin, Z., Ku, S., Lim, T., Park, S. Y., Park, M. S., Ji, G. E., O’Brien, K., & Hwang, K. T. (2021). Antioxidant and Anti-Inflammatory Properties of Recombinant Bifidobacterium bifidum BGN4 Expressing Antioxidant Enzymes. Microorganisms, 9(3), 595. https://doi.org/10.3390/microorganisms9030595






