Genome Sequencing of five Lacticaseibacillus Strains and Analysis of Type I and II Toxin-Antitoxin System Distribution
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Culture Conditions
2.2. Nucleic Acid Extraction, NGS Library Preparation and Sequencing
2.3. Genome Assembly, Annotation and Bioinformatics Analysis
3. Results
3.1. Genomic Features of the Lacticaseibacillus Isolates
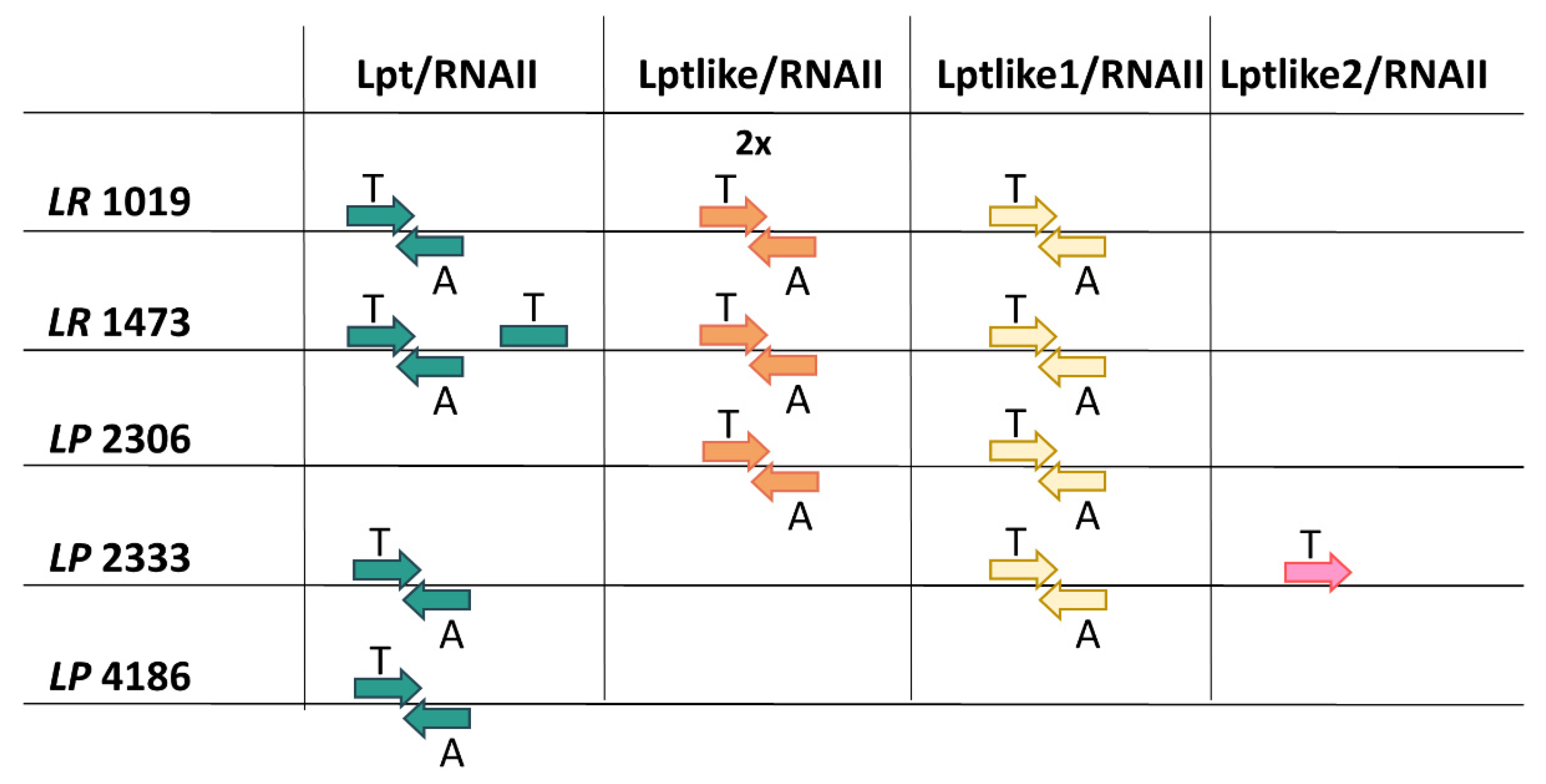
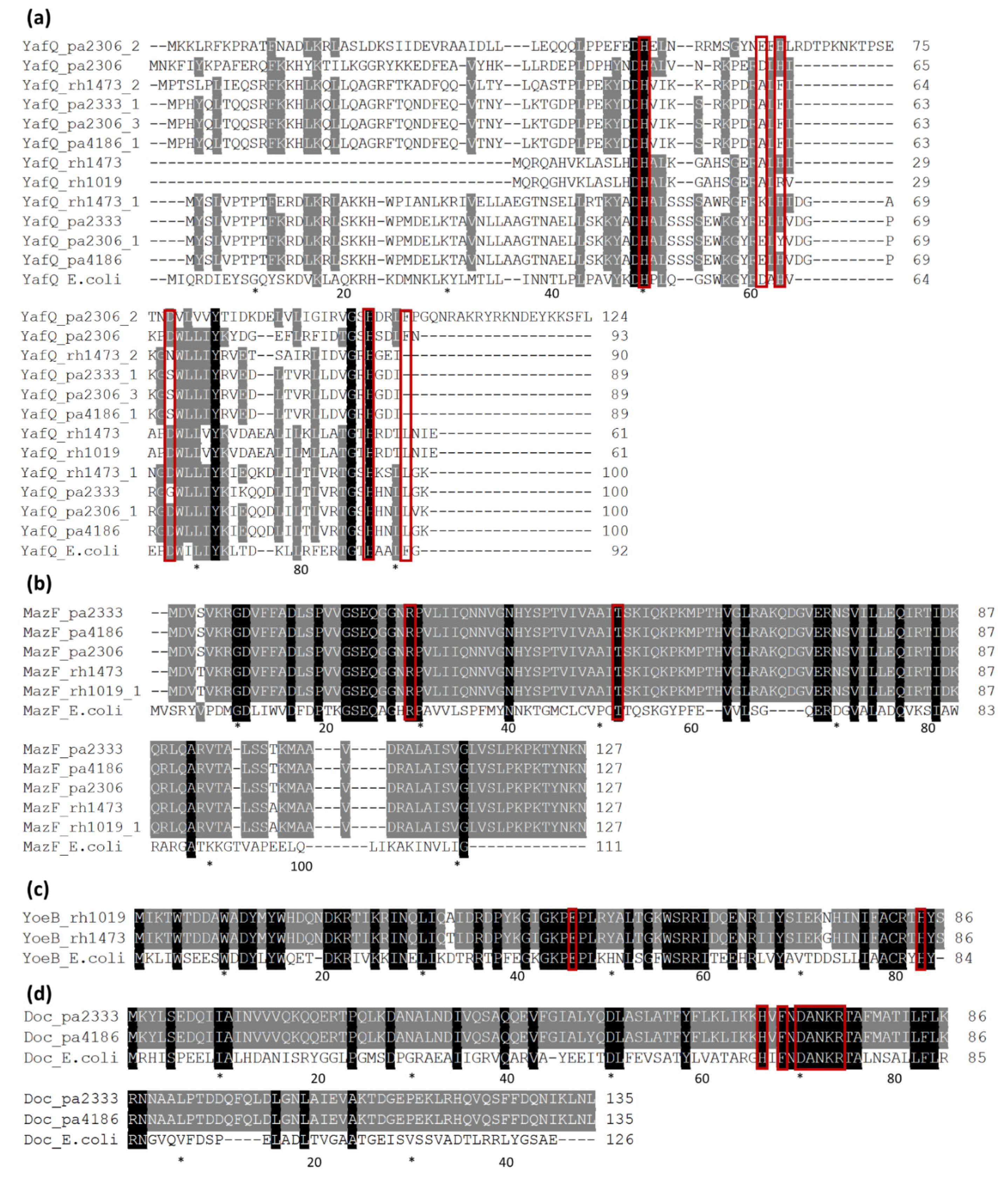
3.2. Type I TA Systems Distribution and Structure
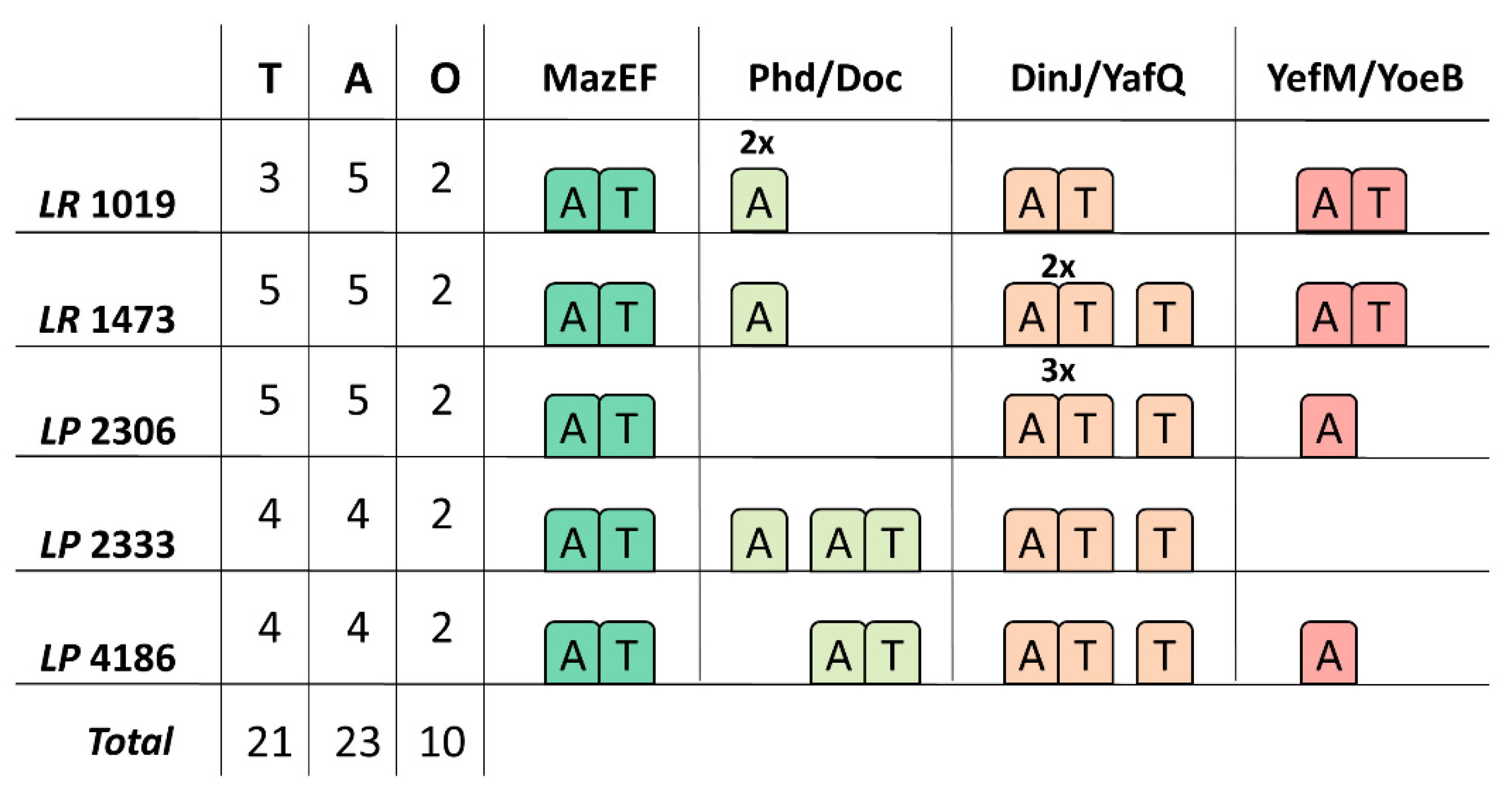
3.3. Type II TA Systems Distribution and Characteristics
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Page, R.; Peti, W. Toxin-antitoxin systems in bacterial growth arrest and persistence. Nat. Chem. Biol. 2016, 12, 208–214. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, Y.; Park, J.H.; Inouye, M. Toxin-antitoxin systems in bacteria and archaea. Annu. Rev. Genet. 2011, 45, 61–79. [Google Scholar] [CrossRef] [PubMed]
- Harms, A.; Brodersen, D.E.; Mitarai, N.; Gerdes, K. Toxins, Targets, and Triggers: An Overview of Toxin-Antitoxin Biology. Mol. Cell 2018, 70, 768–784. [Google Scholar] [CrossRef]
- Kato, F.; Yoshizumi, S.; Yamaguchi, Y.; Inouye, M. Genome-Wide Screening for Identification of Novel Toxin- Antitoxin Systems in Staphylococcus aureus. Appl. Environ. Microbiol. 2019, 85, e00915-19. [Google Scholar] [CrossRef]
- Mohammadzadeh, R.; Shivaee, A.; Ohadi, E.; Kalani, B.S. In Silico Insight into the Dominant Type II Toxin–Antitoxin Systems and Clp Proteases in Listeria monocytogenes and Designation of Derived Peptides as a Novel Approach to Interfere with this System. Int. J. Pept. Res. Ther. 2020, 26, 613–623. [Google Scholar] [CrossRef]
- Broadbent, J.R.; Neeno-Eckwall, E.C.; Stahl, B.; Tandee, K.; Cai, H.; Morovic, W.; Horvath, P.; Heidenreich, J.; Perna, N.T.; Barrangou, R.; et al. Analysis of the Lactobacillus casei supragenome and its influence in species evolution and lifestyle adaptation. BMC Genom. 2012, 13, 533–551. [Google Scholar] [CrossRef]
- Duar, R.M.; Lin, X.B.; Zheng, J.; Martino, M.E.; Grenier, T.; Pérez-Muñoz, M.E.; Leulier, F.; Gänzle, M.; Walter, J. Lifestyles in transition: Evolution and natural history of the genus Lactobacillus. FEMS Microbiol. Rev. 2017, 41, S27–S48. [Google Scholar] [CrossRef]
- Ricci, A.; Levante, A.; Cirlini, M.; Calani, L.; Bernini, V.; Del Rio, D.; Galaverna, G.; Neviani, E.; Lazzi, C. The Influence of Viable Cells and Cell-Free Extracts of Lactobacillus casei on Volatile Compounds and Polyphenolic Profile of Elderberry Juice. Front. Microbiol. 2018, 9, 2784. [Google Scholar] [CrossRef]
- Zheng, J.; Wittouck, S.; Salvetti, E.; Franz, C.M.A.P.; Harris, H.M.B.; Mattarelli, P.; O’toole, P.W.; Pot, B.; Vandamme, P.; Walter, J.; et al. A taxonomic note on the genus Lactobacillus: Description of 23 novel genera, emended description of the genus Lactobacillus beijerinck 1901, and union of Lactobacillaceae and Leuconostocaceae. Int. J. Syst. Evol. Microbiol. 2020, 70, 2782–2858. [Google Scholar] [CrossRef] [PubMed]
- Smokvina, T.; Wels, M.; Polka, J.; Chervaux, C.; Brisse, S.; Boekhorst, J.; van Vlieg, J.E.T.H.; Siezen, R.J. Lactobacillus paracasei Comparative Genomics: Towards Species Pan-Genome Definition and Exploitation of Diversity. PLoS ONE 2013, 8, e68731. [Google Scholar] [CrossRef]
- Toh, H.; Oshima, K.; Nakano, A.; Takahata, M.; Murakami, M.; Takaki, T.; Nishiyama, H.; Igimi, S.; Hattori, M.; Morita, H. Genomic adaptation of the Lactobacillus casei group. PLoS ONE 2013, 8, e75073. [Google Scholar] [CrossRef] [PubMed]
- Douillard, F.P.; Ribbera, A.; Kant, R.; Pietilä, T.E.; Järvinen, H.M.; Messing, M.; Randazzo, C.L.; Paulin, L.; Laine, P.; Ritari, J.; et al. Comparative Genomic and Functional Analysis of 100 Lactobacillus rhamnosus Strains and Their Comparison with Strain GG. PLoS Genet. 2013, 9, e1003683. [Google Scholar] [CrossRef] [PubMed]
- Fontana, A.; Zacconi, C.; Morelli, L. Genetic Signatures of Dairy Lactobacillus casei Group. Front. Microbiol. 2018, 9, 2611. [Google Scholar] [CrossRef]
- Stefanovic, E.; McAuliffe, O. Comparative genomic and metabolic analysis of three Lactobacillus paracasei cheese isolates reveals considerable genomic differences in strains from the same niche. BMC Genom. 2018, 19, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Gobbetti, M.; Di Cagno, R.; Calasso, M.; Neviani, E.; Fox, P.F.; De Angelis, M. Drivers that establish and assembly the lactic acid bacteria biota in cheeses. Trends Food Sci. Technol. 2018, 78, 244–254. [Google Scholar] [CrossRef]
- Levante, A.; Folli, C.; Montanini, B.; Ferrari, A.; Neviani, E.; Lazzi, C. Expression of DinJ-YafQ System of Lactobacillus casei group strains in response to food processing stresses. Microorganisms 2019, 7, 438. [Google Scholar] [CrossRef]
- Folli, C.; Levante, A.; Percudani, R.; Amidani, D.; Bottazzi, S.; Ferrari, A.; Rivetti, C.; Neviani, E.; Lazzi, C. Toward the identification of a type I toxin-antitoxin system in the plasmid DNA of dairy Lactobacillus rhamnosus. Sci. Rep. 2017, 7, 12051. [Google Scholar] [CrossRef]
- Maggi, S.; Yabre, K.; Ferrari, A.; Lazzi, C.; Kawano, M.; Rivetti, C.; Folli, C. Functional characterization of the type I toxin Lpt from Lactobacillus rhamnosus by fluorescence and atomic force microscopy. Sci. Rep. 2019, 9, 1–13. [Google Scholar] [CrossRef]
- Ferrari, A.; Maggi, S.; Montanini, B.; Levante, A.; Lazzi, C.; Yamaguchi, Y.; Rivetti, C.; Folli, C. Identification and first characterization of DinJ-YafQ toxin-antitoxin systems in Lactobacillus species of biotechnological interest. Sci. Rep. 2019, 9, 7645. [Google Scholar] [CrossRef] [PubMed]
- Klimina, K.M.; Kjasova, D.K.; Poluektova, E.U.; Krügel, H.; Leuschner, Y.; Saluz, H.P.; Danilenko, V.N. Identification and characterization of toxin-antitoxin systems in strains of Lactobacillus rhamnosus isolated from humans. Anaerobe 2013, 22, 82–89. [Google Scholar] [CrossRef]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 2017, 13, e1005595. [Google Scholar] [CrossRef]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality assessment tool for genome assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef] [PubMed]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef]
- Page, A.J.; Cummins, C.A.; Hunt, M.; Wong, V.K.; Reuter, S.; Holden, M.T.G.; Fookes, M.; Falush, D.; Keane, J.A.; Parkhill, J. Roary: Rapid large-scale prokaryote pan genome analysis. Bioinformatics 2015, 31, 3691–3693. [Google Scholar] [CrossRef] [PubMed]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Sato, Y.; Morishima, K. BlastKOALA and GhostKOALA: KEGG Tools for Functional Characterization of Genome and Metagenome Sequences. J. Mol. Biol. 2016, 428, 726–731. [Google Scholar] [CrossRef]
- Nicholas, K.B.; Nicholas, H.B.J.; Deerfield, D.W. GeneDoc: Analysis and Visualization of Genetic Variation. EMB News 1997, 4, 14. [Google Scholar]
- Savo Sardaro, M.L.; Levante, A.; Bernini, V.; Gatti, M.; Neviani, E.; Lazzi, C. The spxB gene as a target to identify Lactobacillus casei group species in cheese. Food Microbiol. 2016, 59, 57–65. [Google Scholar] [CrossRef]
- Maehigashi, T.; Ruangprasert, A.; Miles, S.J.; Dunham, C.M. Molecular basis of ribosome recognition and mRNA hydrolysis by the E. coli YafQ toxin. Nucleic Acids Res. 2015, 43, 8002–8012. [Google Scholar] [CrossRef]
- Simanshu, D.; Yamaguchi, Y.; Park, J.-H.; Inouye, M.; Patel, D. Structural Basis of mRNA Recognition and Cleavage by Toxin MazF and Its Regulation by Antitoxin MazE in Bacillus subtilis. Mol. Cell 2013, 52, 447–458. [Google Scholar] [CrossRef]
- Feng, S.; Chen, Y.; Kamada, K.; Wang, H.; Tang, K.; Wang, M.; Gao, Y.G. YoeB-ribosome structure: A canonical RNase that requires the ribosome for its specific activity. Nucleic Acids Res. 2013, 41, 9549–9556. [Google Scholar] [CrossRef] [PubMed]
- Bottari, B.; Levante, A.; Neviani, E.; Gatti, M. How the fewest become the greatest. L. casei’s impact on long ripened cheeses. Front. Microbiol. 2018, 9, 2866. [Google Scholar] [PubMed]
- Van Mastrigt, O.; Abee, T.; Lillevang, S.K.; Smid, E.J. Quantitative physiology and aroma formation of a dairy Lactococcus lactis at near-zero growth rates. Food Microbiol. 2018, 73, 216–226. [Google Scholar] [CrossRef] [PubMed]
- Davray, D.; Deo, D.; Kulkarni, R. Plasmids encode niche-specific traits in Lactobacillaceae. Microb. Genom. 2020. [Google Scholar] [CrossRef]
- Bancalari, E.; Montanari, C.; Levante, A.; Alinovi, M.; Neviani, E.; Gardini, F.; Gatti, M. Lactobacillus paracasei 4341 as adjunct culture to enhance flavor in short ripened Caciotta-type cheese. Food Res. Int. 2020, 135, 109284. [Google Scholar] [CrossRef]
- Wüthrich, D.; Wenzel, C.; Bavan, T.; Bruggmann, R.; Berthoud, H.; Irmler, S. Transcriptional regulation of cysteine and methionine metabolism in Lactobacillus paracasei FAM18149. Front. Microbiol. 2018, 9, 1261. [Google Scholar] [CrossRef]
- Liu, M.; Prakash, C.; Nauta, A.; Siezen, R.J.; Francke, C. Computational analysis of cysteine and methionine metabolism and its regulation in dairy starter and related bacteria. J. Bacteriol. 2012, 194, 3522–3533. [Google Scholar] [CrossRef]
- Bottari, B.; Levante, A.; Bancalari, E.; Sforza, S.; Bottesini, C.; Prandi, B.; De Filippis, F.; Ercolini, D.; Nocetti, M.; Gatti, M. The Interrelationship Between Microbiota and Peptides During Ripening as a Driver for Parmigiano Reggiano Cheese Quality. Front. Microbiol. 2020, 11, 581658. [Google Scholar] [CrossRef]
- Levante, A.; De Filippis, F.; La Storia, A.; Gatti, M.; Neviani, E.; Ercolini, D.; Lazzi, C. Metabolic gene-targeted monitoring of non-starter lactic acid bacteria during cheese ripening. Int. J. Food Microbiol. 2017, 257, 276–284. [Google Scholar] [CrossRef]
- Riffaud, C.; Pinel-Marie, M.L.; Felden, B. Cross-Regulations between Bacterial Toxin–Antitoxin Systems: Evidence of an Interconnected Regulatory Network? Trends Microbiol. 2020, 28, 851–866. [Google Scholar] [CrossRef]
- Xia, K.; Han, C.; Xu, J.; Liang, X. Transcriptome response of Acetobacter pasteurianus Ab3 to high acetic acid stress during vinegar production. Appl. Microbiol. Biotechnol. 2020, 104, 10585–10599. [Google Scholar] [CrossRef] [PubMed]
- Levante, A.; Bancalari, E.; Tambassi, M.; Lazzi, C.; Neviani, E. Phenotypic Diversity of Lactobacillus casei group isolates as a selection criterion for use as secondary adjunct starters. Microorganisms 2020, 8, 128. [Google Scholar] [CrossRef] [PubMed]
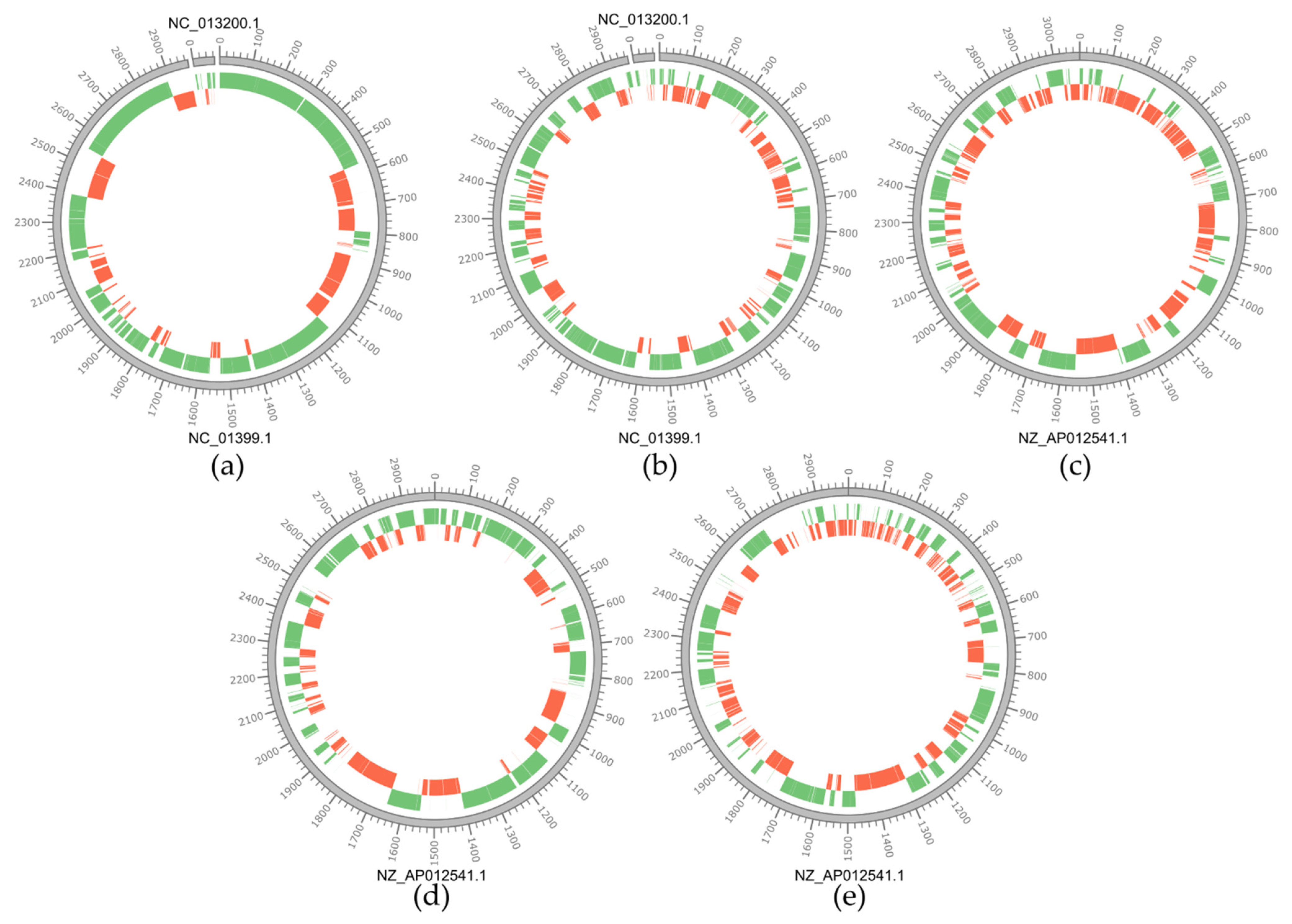
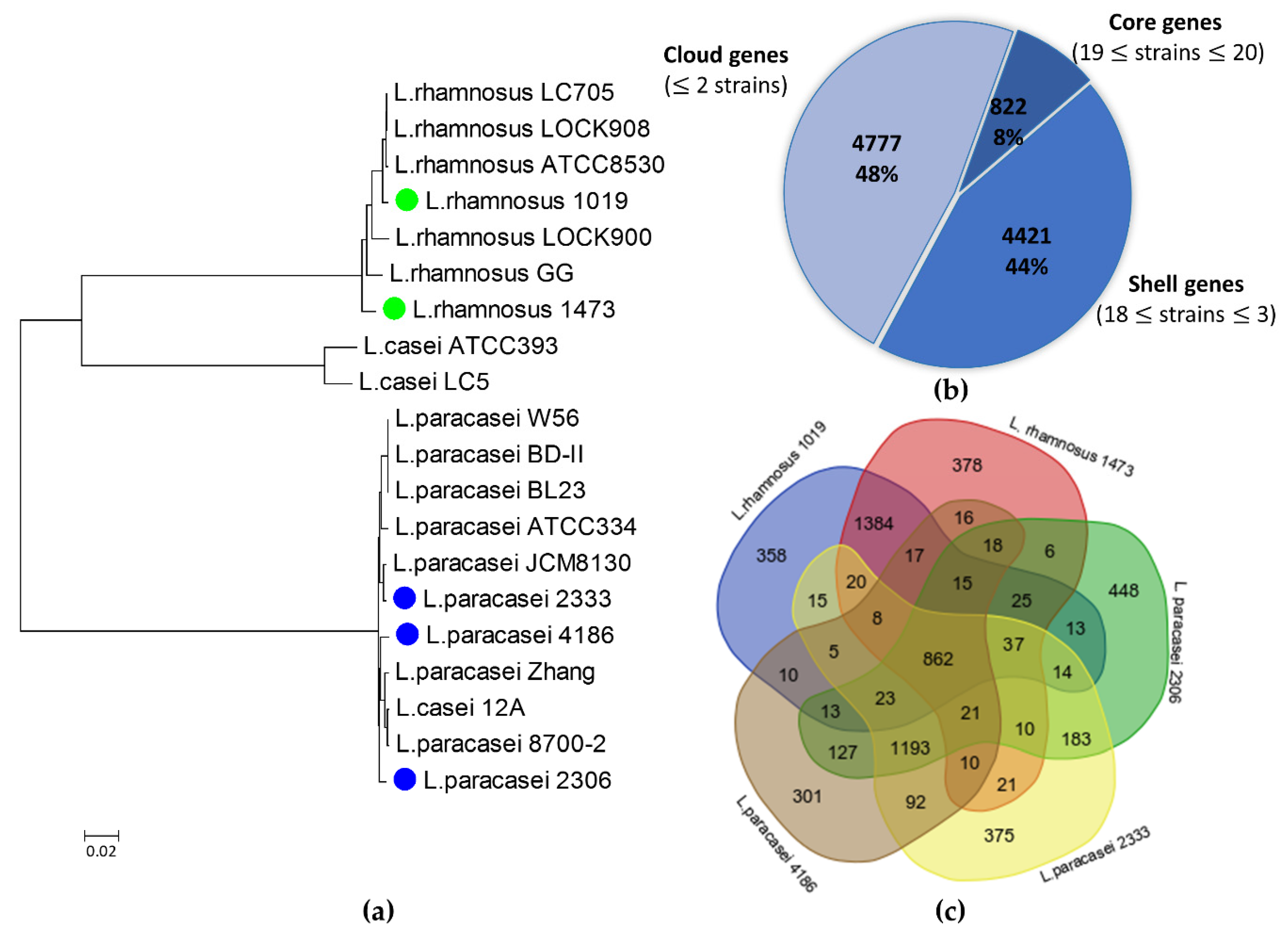

| Strain | Isolation Source 1 | Ripening (months) | Size (bp) | GC Content (%) | n° of Contigs | n° of ORF | cds | ncRNAs |
|---|---|---|---|---|---|---|---|---|
| L. rhamnosus 1019 | PR | 4 | 3,088,347 | 46.58 | 248 | 3300 | 3232 | 68 |
| L. rhamnosus 1473 | PR | 20 | 3,088,357 | 46.57 | 384 | 3464 | 3402 | 62 |
| L. paracasei 2306 | PR | 6 | 3,240,492 | 46.12 | 364 | 3637 | 3572 | 65 |
| L. paracasei 2333 | PT | n.a. | 3,163,164 | 46.54 | 993 | 3979 | 3915 | 64 |
| L. paracasei 4186 | PR | 1 | 2,922,588 | 46.27 | 396 | 3326 | 3264 | 62 |
| Name | Length (bp) | Coverage (x) | mL | pf | ta | Plasmid |
|---|---|---|---|---|---|---|
| p1019-1 | 27,278 | 185 | x | x | CP014202.1 | |
| p1019-2 | 4812 | 1487 | x | x | CP001155.1 | |
| p1019-3 | 3955 | 177 | x | CP048004.1 | ||
| p1019-4 | 3644 | 1241 | x | AY662331.1 | ||
| p1019-5 | 2422 | 947 | x | AP012543.1 | ||
| p1473-1 | 15,540 | 584 | xx | CP012190.1 | ||
| p1473-2 | 14,433 | 121 | x | x | CP001156.1 | |
| p1473-3 | 13,211 | 131 | x | CP044229.1 | ||
| p1473-4 | 8964 | 623 | x | x | EU255257.1 | |
| p1473-5 | 5983 | 25 | x | MH621333.1 | ||
| p2306-1 | 10,022 | 164 | x | x | CP044229.1 | |
| p2306-2 | 24,039 | 187 | x | CP035564.1 | ||
| p2306-3 | 8837 | 937 | x | AY662330.1 | ||
| p2306-4 | 5006 | 1682 | x | EU255257.1 | ||
| p2306-5 | 3223 | 141 | x | CP048004.1 | ||
| p2333-1 | 23,737 | 179 | x | CP014885.1 | ||
| p2333-2 | 11,765 | 709 | x | CP035564.1 | ||
| p2333-3 | 9153 | 618 | x | Z50861.1 | ||
| p2333-4 | 8630 | 191 | x | CP044229.1 | ||
| p2333-5 | 8002 | 615 | x | x | AY662330.1 | |
| p2333-6 | 5455 | 95 | x | CP035564.1 | ||
| p2333-7 | 3897 | 150 | x | CP007125.1 | ||
| p2333-8 | 2076 | 160 | x | x | CP044362.1 | |
| p4186-1 | 5673 | 30 | x | CP007125.1 | ||
| p4186-2 | 3824 | 114 | x | CP012190.1 | ||
| p4186-3 | 3097 | 153 | x | CP017262.1 |
| Strains | Lpt Toxins and Its Homologs | Location |
|---|---|---|
| L. rhamnosus 1019 | Lpt_rh1019 | p1019-5 |
| Lptlike_rh1019 | p1019-2 | |
| Lptlike_rh1019_1 | n.a. | |
| Lptlike1_rh1019 | n.a. | |
| L. rhamnosus 1473 | Lpt_rh1473 | p1473-5 |
| Lpt_rh1473_1 | p1473-1 | |
| Lptlike_rh1473 | p1473-4 | |
| Lptlike1_rh1473 | p1473-1 | |
| L. paracasei 2306 | Lptlike_pa2306 | p2306-4 |
| Lptlike1_pa2306 | p2306-3 | |
| L. paracasei 2333 | Lpt_pa2333 | p2333-3 |
| Lptlike1_pa2333 | p2333-5 | |
| Lptlike2_pa2333 | n.a. | |
| L. paracasei 4186 | Lpt_pa4186 | n.a. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Levante, A.; Lazzi, C.; Vatsellas, G.; Chatzopoulos, D.; Dionellis, V.S.; Makrythanasis, P.; Neviani, E.; Folli, C. Genome Sequencing of five Lacticaseibacillus Strains and Analysis of Type I and II Toxin-Antitoxin System Distribution. Microorganisms 2021, 9, 648. https://doi.org/10.3390/microorganisms9030648
Levante A, Lazzi C, Vatsellas G, Chatzopoulos D, Dionellis VS, Makrythanasis P, Neviani E, Folli C. Genome Sequencing of five Lacticaseibacillus Strains and Analysis of Type I and II Toxin-Antitoxin System Distribution. Microorganisms. 2021; 9(3):648. https://doi.org/10.3390/microorganisms9030648
Chicago/Turabian StyleLevante, Alessia, Camilla Lazzi, Giannis Vatsellas, Dimitris Chatzopoulos, Vasilis S. Dionellis, Periklis Makrythanasis, Erasmo Neviani, and Claudia Folli. 2021. "Genome Sequencing of five Lacticaseibacillus Strains and Analysis of Type I and II Toxin-Antitoxin System Distribution" Microorganisms 9, no. 3: 648. https://doi.org/10.3390/microorganisms9030648
APA StyleLevante, A., Lazzi, C., Vatsellas, G., Chatzopoulos, D., Dionellis, V. S., Makrythanasis, P., Neviani, E., & Folli, C. (2021). Genome Sequencing of five Lacticaseibacillus Strains and Analysis of Type I and II Toxin-Antitoxin System Distribution. Microorganisms, 9(3), 648. https://doi.org/10.3390/microorganisms9030648






