Variability in Bacteriophage and Antibiotic Sensitivity in Serial Pseudomonas aeruginosa Isolates from Cystic Fibrosis Airway Cultures over 12 Months
Abstract
:1. Introduction
2. Materials and Methods
2.1. Pseudomonas aeruginosa Isolates
2.2. Antibiotic Sensitivity Testing
2.3. Bacteriophages and Plaque Assays
2.4. Longitudinal Variability in Antibiotic and Phage Susceptibility
2.5. Variable Number Tandem Repeat (VNTR) Typing
2.6. Lung Function Tests
2.7. Statistical Analysis and Graphical Representation
3. Results
3.1. Patient Demographics and Isolate Characteristics
3.2. Antibiogram Profiles and Phage Susceptibility
| N patients | ||||
|---|---|---|---|---|
| Antimicrobial | Stably Sensitive | Variable | Stably Resistant | Stable/Variable (%) |
| Bacteriophage (neat cocktail) | 10 (*1) | 10 (*2) | 1 | 52/48 (*33/67) |
| Bacteriophage (10−3 dilution) | 1 | 17 (*3) | 3 | 19/81 (*0/100) |
| Amikacin | 6 | 9 (*2) | 8 (*1) | 67/33 (*33/67) |
| Aztreonam | 8(*1) | 6 (*2) | 7 | 71/29 (*33/67) |
| Ceftazidime | 9 | 6 (*3) | 6 | 71/29 (*0/100) |
| Ciprofloxacin | 1 | 9 (*3) | 11 | 57/43 (*0/100) |
| Gentamicin | 4 | 9 (*2* | 8 (*1) | 57/43 (*33/67) |
| Meropenem | 8 | 6 (*2) | 7 (*1) | 71/29 (*33/67) |
| Piperacillin-Tazobactam | 10 | 6 (*2) | 5 (*1) | 71/29 (*33/67) |
| Tobramycin | 14 (*1) | 3 (*2) | 4 | 86/14% (*33/67) |
3.3. Similarity of Results in Patients Sharing a Common Pa Strain
 , with subsequent isolates
, with subsequent isolates  ,
,  ,
,  ,
,  , if there were 5 isolates through the course of 2016. Open symbols represent different strains, identified by VNTR analysis. Abbreviations—AMK: amikacin; AZM: aztreonam; CEF: ceftazidime; CIP: ciprofloxacin; GEN: gentamicin; MER: meropenem; PTZ: piperacillin-tazobactam; SXT: co-trimoxazole (no EUCAST breakpoint); TOB: tobramycin. Disc diffusion assay for antibiotic sensitivity testing was performed once at each time point and EUCAST sensitivity cutoffs are indicated by the bars. Biological triplicates of phage assay are represented individually and bars indicate both neat phage (>2 × 102 PFU/mL) and dilute phage (>2 × 105 PFU/mL) cutoffs. (a–d) Patients G, K, M and N were colonized with the LES. All strains qualified as MDR based on antibiograms and 14/18 (78%) LES isolates showed some lytic activity to phage using the lower sensitivity cutoff. Using the higher sensitivity cutoff of 103 serial dilutions, 9/18 (50%) showed sensitivity to phage. (a) Patient G had isolates that are broadly resistant to antibiotics-sensitive to tobramycin at 2 and to meropenem at 1 time points, respectively, but broadly sensitive to phage at all time points; (b) Patient K was colonized by a different strain at the first time point, but all subsequent isolates were LES with lytic activity to phage; (c) Patient M’s LES strain was sensitive only to piperacillin-tazobactam but sensitive to phage at all time points; and (d) Patient N was colonized with LES resistant to all antibiotics tested. Only 1 of this patient’s 5 isolates showed lytic activity to the neat phage cocktail and no isolates were deemed sensitive using the higher cutoff.
, if there were 5 isolates through the course of 2016. Open symbols represent different strains, identified by VNTR analysis. Abbreviations—AMK: amikacin; AZM: aztreonam; CEF: ceftazidime; CIP: ciprofloxacin; GEN: gentamicin; MER: meropenem; PTZ: piperacillin-tazobactam; SXT: co-trimoxazole (no EUCAST breakpoint); TOB: tobramycin. Disc diffusion assay for antibiotic sensitivity testing was performed once at each time point and EUCAST sensitivity cutoffs are indicated by the bars. Biological triplicates of phage assay are represented individually and bars indicate both neat phage (>2 × 102 PFU/mL) and dilute phage (>2 × 105 PFU/mL) cutoffs. (a–d) Patients G, K, M and N were colonized with the LES. All strains qualified as MDR based on antibiograms and 14/18 (78%) LES isolates showed some lytic activity to phage using the lower sensitivity cutoff. Using the higher sensitivity cutoff of 103 serial dilutions, 9/18 (50%) showed sensitivity to phage. (a) Patient G had isolates that are broadly resistant to antibiotics-sensitive to tobramycin at 2 and to meropenem at 1 time points, respectively, but broadly sensitive to phage at all time points; (b) Patient K was colonized by a different strain at the first time point, but all subsequent isolates were LES with lytic activity to phage; (c) Patient M’s LES strain was sensitive only to piperacillin-tazobactam but sensitive to phage at all time points; and (d) Patient N was colonized with LES resistant to all antibiotics tested. Only 1 of this patient’s 5 isolates showed lytic activity to the neat phage cocktail and no isolates were deemed sensitive using the higher cutoff.
 , with subsequent isolates
, with subsequent isolates  ,
,  ,
,  ,
,  , if there were 5 isolates through the course of 2016. Open symbols represent different strains, identified by VNTR analysis. Abbreviations—AMK: amikacin; AZM: aztreonam; CEF: ceftazidime; CIP: ciprofloxacin; GEN: gentamicin; MER: meropenem; PTZ: piperacillin-tazobactam; SXT: co-trimoxazole (no EUCAST breakpoint); TOB: tobramycin. Disc diffusion assay for antibiotic sensitivity testing was performed once at each time point and EUCAST sensitivity cutoffs are indicated by the bars. Biological triplicates of phage assay are represented individually and bars indicate both neat phage (>2 × 102 PFU/mL) and dilute phage (>2 × 105 PFU/mL) cutoffs. (a–d) Patients G, K, M and N were colonized with the LES. All strains qualified as MDR based on antibiograms and 14/18 (78%) LES isolates showed some lytic activity to phage using the lower sensitivity cutoff. Using the higher sensitivity cutoff of 103 serial dilutions, 9/18 (50%) showed sensitivity to phage. (a) Patient G had isolates that are broadly resistant to antibiotics-sensitive to tobramycin at 2 and to meropenem at 1 time points, respectively, but broadly sensitive to phage at all time points; (b) Patient K was colonized by a different strain at the first time point, but all subsequent isolates were LES with lytic activity to phage; (c) Patient M’s LES strain was sensitive only to piperacillin-tazobactam but sensitive to phage at all time points; and (d) Patient N was colonized with LES resistant to all antibiotics tested. Only 1 of this patient’s 5 isolates showed lytic activity to the neat phage cocktail and no isolates were deemed sensitive using the higher cutoff.
, if there were 5 isolates through the course of 2016. Open symbols represent different strains, identified by VNTR analysis. Abbreviations—AMK: amikacin; AZM: aztreonam; CEF: ceftazidime; CIP: ciprofloxacin; GEN: gentamicin; MER: meropenem; PTZ: piperacillin-tazobactam; SXT: co-trimoxazole (no EUCAST breakpoint); TOB: tobramycin. Disc diffusion assay for antibiotic sensitivity testing was performed once at each time point and EUCAST sensitivity cutoffs are indicated by the bars. Biological triplicates of phage assay are represented individually and bars indicate both neat phage (>2 × 102 PFU/mL) and dilute phage (>2 × 105 PFU/mL) cutoffs. (a–d) Patients G, K, M and N were colonized with the LES. All strains qualified as MDR based on antibiograms and 14/18 (78%) LES isolates showed some lytic activity to phage using the lower sensitivity cutoff. Using the higher sensitivity cutoff of 103 serial dilutions, 9/18 (50%) showed sensitivity to phage. (a) Patient G had isolates that are broadly resistant to antibiotics-sensitive to tobramycin at 2 and to meropenem at 1 time points, respectively, but broadly sensitive to phage at all time points; (b) Patient K was colonized by a different strain at the first time point, but all subsequent isolates were LES with lytic activity to phage; (c) Patient M’s LES strain was sensitive only to piperacillin-tazobactam but sensitive to phage at all time points; and (d) Patient N was colonized with LES resistant to all antibiotics tested. Only 1 of this patient’s 5 isolates showed lytic activity to the neat phage cocktail and no isolates were deemed sensitive using the higher cutoff.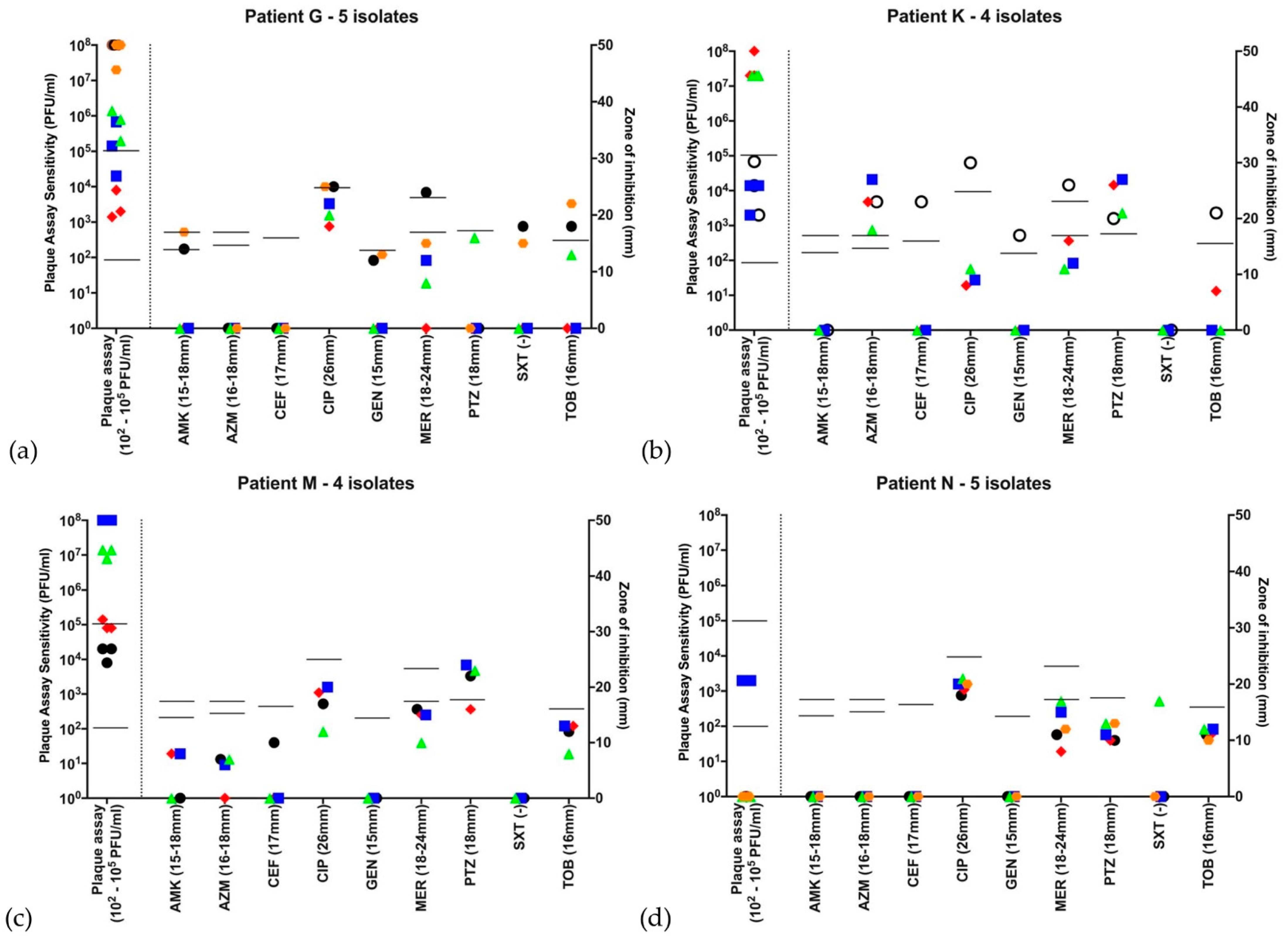
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cystic Fibrosis Strength in Numbers; UK Cystic Fibrosis Registry Annual Data Report 2018; Cystic Fibrosis Trust: London, UK, 2019; Available online: https://www.cysticfibrosis.org.uk/sites/default/files/2020-12/2018%20Registry%20Annual%20Data%20Report.pdf (accessed on 19 March 2021).
- Hoiby, N.; Frederiksen, B.; Pressler, T. Eradication of early Pseudomonas aeruginosa infection. J. Cyst Fibros 2005, 4, 49–54. [Google Scholar] [CrossRef] [Green Version]
- Burns, J.L.; Gibson, R.L.; McNamara, S.; Yim, D.; Emerson, J.; Rosenfeld, M.; Hiatt, P.; McCoy, K.; Castile, R.; Smith, A.L.; et al. Longitudinal assessment of Pseudomonas aeruginosa in young children with cystic fibrosis. J. Infect. Dis. 2001, 183, 444–452. [Google Scholar] [CrossRef] [Green Version]
- Tümmler, B.; Koopmann, U.; Grothues, D.; Weissbrodt, H.; Steinkamp, G.; von der Hardt, H. Nosocomial Aqcuisition of Pseudomonas aeruginosa by Cystic Fibrosis Patients. J. Med Microbiol. 1991, 29, 1265–1267. [Google Scholar]
- Aloush, V.; Navon-Venezia, S.; Seigman-Igra, Y.; Cabili, S.; Carmeli, Y. Multidrug-resistant Pseudomonas aeruginosa: Risk factors and clinical impact. Antimicrob. Agents Chemother. 2006, 50, 43–48. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, K.; Smyth, R.L.; Govan, J.R.; Doherty, C.; Winstanley, C.; Denning, N.; Heaf, D.P.; van Saene, H.; Hart, C.A. Spread of beta-lactam-resistant Pseudomonas aeruginosa in a cystic fibrosis clinic. Lancet 1996, 348, 639–642. [Google Scholar] [CrossRef]
- Langton Hewer, S.C.; Smyth, A.R. Antibiotic strategies for eradicating Pseudomonas aeruginosa in people with cystic fibrosis. Cochrane Database Syst Rev. 2014, 11, CD004197. [Google Scholar] [CrossRef] [Green Version]
- Hansen, C.R.; Pressler, T.; Hoiby, N. Early aggressive eradication therapy for intermittent Pseudomonas aeruginosa airway colonization in cystic fibrosis patients: 15 years experience. J. Cyst. Fibros. 2008, 7, 523–530. [Google Scholar] [CrossRef] [Green Version]
- Folkesson, A.; Jelsbak, L.; Yang, L.; Johansen, H.K.; Ciofu, O.; Hoiby, N.; Molin, S. Adaptation of Pseudomonas aeruginosa to the cystic fibrosis airway: An evolutionary perspective. Nat. Rev. Microbiol. 2012, 10, 841–851. [Google Scholar] [CrossRef]
- Mowat, E.; Paterson, S.; Fothergill, J.L.; Wright, E.A.; Ledson, M.J.; Walshaw, M.J.; Brockhurst, M.A.; Winstanley, C. Pseudomonas aeruginosa population diversity and turnover in cystic fibrosis chronic infections. Am. J. Respir. Crit. Care Med. 2011, 183, 1674–1679. [Google Scholar] [CrossRef]
- Fernandez-Barat, L.; Ciofu, O.; Kragh, K.N.; Pressler, T.; Johansen, U.; Motos, A.; Torres, A.; Hoiby, N. Phenotypic shift in Pseudomonas aeruginosa populations from cystic fibrosis lungs after 2-week antipseudomonal treatment. J. Cyst. Fibros. 2017, 16, 222–229. [Google Scholar] [CrossRef] [Green Version]
- Doring, G.; Flume, P.; Heijerman, H.; Elborn, J.S.; Consensus Study, G. Treatment of lung infection in patients with cystic fibrosis: Current and future strategies. J. Cyst. Fibros. 2012, 11, 461–479. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pitt, T.L.; Sparrow, M.; Warner, M.; Stefanidou, M. Survey of resistance of Pseudomonas aeruginosa from UK patients with cystic fibrosis to six commonly prescribed antimicrobial agents. Thorax 2003, 58, 794–796. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hirsch, E.B.; Tam, V.H. Impact of multidrug-resistant Pseudomonas aeruginosa infection on patient outcomes. Expert Rev. Pharmacoecon. Outcomes Res. 2010, 10, 441–451. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lucca, F.; Guarnieri, M.; Ros, M.; Muffato, G.; Rigoli, R.; Da Dalt, L. Antibiotic resistance evolution of Pseudomonas aeruginosa in cystic fibrosis patients (2010–2013). Clin. Respir. J. 2018, 12, 2189–2196. [Google Scholar] [CrossRef] [PubMed]
- Hurley, M.N.; Ariff, A.H.; Bertenshaw, C.; Bhatt, J.; Smyth, A.R. Results of antibiotic susceptibility testing do not influence clinical outcome in children with cystic fibrosis. J. Cyst. Fibros. 2012, 11, 288–292. [Google Scholar] [CrossRef] [Green Version]
- Smith, A.L.; Fiel, S.B.; Mayer-Hamblett, N.; Ramsey, B.; Burns, J.L. Susceptibility testing of Pseudomonas aeruginosa isolates and clinical response to parenteral antibiotic administration: Lack of association in cystic fibrosis. Chest 2003, 123, 1495–1502. [Google Scholar] [CrossRef] [Green Version]
- Fong, S.A.; Drilling, A.; Morales, S.; Cornet, M.E.; Woodworth, B.A.; Fokkens, W.J.; Psaltis, A.J.; Vreugde, S.; Wormald, P.J. Activity of Bacteriophages in Removing Biofilms of Pseudomonas aeruginosa Isolates from Chronic Rhinosinusitis Patients. Front. Cell. Infect. Microbiol. 2017, 7, 418. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dedrick, R.M.; Guerrero-Bustamante, C.A.; Garlena, R.A.; Russell, D.A.; Ford, K.; Harris, K.; Gilmour, K.C.; Soothill, J.; Jacobs-Sera, D.; Schooley, R.T.; et al. Engineered bacteriophages for treatment of a patient with a disseminated drug-resistant Mycobacterium abscessus. Nat. Med. 2019, 25, 730–733. [Google Scholar] [CrossRef]
- Pabary, R.; Singh, C.; Morales, S.; Bush, A.; Alshafi, K.; Bilton, D.; Alton, E.W.; Smithyman, A.; Davies, J.C. Antipseudomonal Bacteriophage Reduces Infective Burden and Inflammatory Response in Murine Lung. Antimicrob. Agents Chemother. 2016, 60, 744–751. [Google Scholar] [CrossRef] [Green Version]
- Law, N.; Logan, C.; Yung, G.; Furr, C.L.; Lehman, S.M.; Morales, S.; Rosas, F.; Gaidamaka, A.; Bilinsky, I.; Grint, P.; et al. Successful adjunctive use of bacteriophage therapy for treatment of multidrug-resistant Pseudomonas aeruginosa infection in a cystic fibrosis patient. Infection 2019, 47, 665–668. [Google Scholar] [CrossRef] [PubMed]
- Pusch, W.; Kostrzewa, M. Application of MALDI-TOF mass spectrometry in screening and diagnostic research. Curr. Pharm. Des. 2005, 11, 2577–2591. [Google Scholar] [CrossRef] [PubMed]
- Matuschek, E.; Brown, D.F.; Kahlmeter, G. Development of the EUCAST disk diffusion antimicrobial susceptibility testing method and its implementation in routine microbiology laboratories. Clin. Microbiol. Infect. 2014, 20, O255–O266. [Google Scholar] [CrossRef] [Green Version]
- Magiorakos, A.P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef] [Green Version]
- Aslam, S.; Courtwright, A.M.; Koval, C.; Lehman, S.M.; Morales, S.; Furr, C.L.; Rosas, F.; Brownstein, M.J.; Fackler, J.R.; Sisson, B.M.; et al. Early clinical experience of bacteriophage therapy in 3 lung transplant recipients. Am. J. Transplant. 2019, 19, 2631–2639. [Google Scholar] [CrossRef] [PubMed]
- Dong-ju, K. Relation of microbial biomass to counting units for Pseudomonas aeruginosa. Afr. J. Microbiol. Res. 2012, 6, 4620–4622. [Google Scholar] [CrossRef]
- Turton, J.F.; Turton, S.E.; Yearwood, L.; Yarde, S.; Kaufmann, M.E.; Pitt, T.L. Evaluation of a nine-locus variable-number tandem-repeat scheme for typing of Pseudomonas aeruginosa. Clin. Microbiol. Infect. 2010, 16, 1111–1116. [Google Scholar] [CrossRef] [Green Version]
- Quanjer, P. Standardized lung function testing. ReportWorking Party Standardization of Lung Function Tests. European Community for Coal and Steel. Bull. Eur. Physiopathol. Resp. 1983, 19, 1–95. [Google Scholar]
- Rosenthal, M.; Bain, S.H.; Cramer, D.; Helms, P.; Denison, D.; Bush, A.; Warner, J.O. Lung function in white children aged 4 to 19 years: I—Spirometry. Thorax 1993, 48, 794–802. [Google Scholar] [CrossRef] [Green Version]
- Martin, K.; Baddal, B.; Mustafa, N.; Perry, C.; Underwood, A.; Constantidou, C.; Loman, N.; Kenna, D.T.; Turton, J.F. Clusters of genetically similar isolates of Pseudomonas aeruginosa from multiple hospitals in the UK. J. Med. Microbiol. 2013, 62, 988–1000. [Google Scholar] [CrossRef] [Green Version]
- Jeukens, J.; Boyle, B.; Kukavica-Ibrulj, I.; Ouellet, M.M.; Aaron, S.D.; Charette, S.J.; Fothergill, J.L.; Tucker, N.P.; Winstanley, C.; Levesque, R.C. Comparative genomics of isolates of a Pseudomonas aeruginosa epidemic strain associated with chronic lung infections of cystic fibrosis patients. PLoS ONE 2014, 9, e87611. [Google Scholar] [CrossRef] [Green Version]
- Abedon, S.T. Phage-Antibiotic Combination Treatments: Antagonistic Impacts of Antibiotics on the Pharmacodynamics of Phage Therapy? Antibiotics 2019, 8, 182. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tagliaferri, T.L.; Jansen, M.; Horz, H.P. Fighting Pathogenic Bacteria on Two Fronts: Phages and Antibiotics as Combined Strategy. Front. Cell. Infect. Microbiol. 2019, 9, 22. [Google Scholar] [CrossRef] [PubMed]
- Willner, D.; Haynes, M.R.; Furlan, M.; Schmieder, R.; Lim, Y.W.; Rainey, P.B.; Rohwer, F.; Conrad, D. Spatial distribution of microbial communities in the cystic fibrosis lung. ISME J. 2012, 6, 471–474. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cramer, N.; Wiehlmann, L.; Ciofu, O.; Tamm, S.; Hoiby, N.; Tummler, B. Molecular epidemiology of chronic Pseudomonas aeruginosa airway infections in cystic fibrosis. PLoS ONE 2012, 7, e50731. [Google Scholar] [CrossRef]
- Foweraker, J.E.; Laughton, C.R.; Brown, D.F.; Bilton, D. Phenotypic variability of Pseudomonas aeruginosa in sputa from patients with acute infective exacerbation of cystic fibrosis and its impact on the validity of antimicrobial susceptibility testing. J. Antimicrob. Chemother. 2005, 55, 921–927. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fyfe, J.A.; Govan, J.R. Chromosomal loci associated with antibiotic hypersensitivity in pulmonary isolates of Pseudomonas aeruginosa. J. Gen. Microbiol. 1984, 130, 825–834. [Google Scholar] [CrossRef] [Green Version]
- Wong, A.; Kassen, R. Parallel evolution and local differentiation in quinolone resistance in Pseudomonas aeruginosa. Microbiology 2011, 157, 937–944. [Google Scholar] [CrossRef]
- Labrie, S.J.; Samson, J.E.; Moineau, S. Bacteriophage resistance mechanisms. Nat. Rev. Microbiol. 2010, 8, 317–327. [Google Scholar] [CrossRef]
- Davies, E.V.; James, C.E.; Kukavica-Ibrulj, I.; Levesque, R.C.; Brockhurst, M.A.; Winstanley, C. Temperate phages enhance pathogen fitness in chronic lung infection. ISME J. 2016, 10, 2553–2555. [Google Scholar] [CrossRef]
- Rice, S.A.; Tan, C.H.; Mikkelsen, P.J.; Kung, V.; Woo, J.; Tay, M.; Hauser, A.; McDougald, D.; Webb, J.S.; Kjelleberg, S. The biofilm life cycle and virulence of Pseudomonas aeruginosa are dependent on a filamentous prophage. ISME J. 2009, 3, 271–282. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ambroa, A.; Blasco, L.; Lopez-Causape, C.; Trastoy, R.; Fernandez-Garcia, L.; Bleriot, I.; Ponce-Alonso, M.; Pacios, O.; Lopez, M.; Canton, R.; et al. Temperate Bacteriophages (Prophages) in Pseudomonas aeruginosa Isolates Belonging to the International Cystic Fibrosis Clone (CC274). Front. Microbiol. 2020, 11, 556706. [Google Scholar] [CrossRef] [PubMed]
- Davies, J.C.; Martin, I. New anti-pseudomonal agents for cystic fibrosis- still needed in the era of small molecule CFTR modulators? Expert Opin. Pharmacother. 2018, 19, 1327–1336. [Google Scholar] [CrossRef] [PubMed]
- Hisert, K.B.; Heltshe, S.L.; Pope, C.; Jorth, P.; Wu, X.; Edwards, R.M.; Radey, M.; Accurso, F.J.; Wolter, D.J.; Cooke, G.; et al. Restoring Cystic Fibrosis Transmembrane Conductance Regulator Function Reduces Airway Bacteria and Inflammation in People with Cystic Fibrosis and Chronic Lung Infections. Am. J. Respir. Crit. Care Med. 2017, 195, 1617–1628. [Google Scholar] [CrossRef] [PubMed]
 , with subsequent isolates
, with subsequent isolates  ,
,  ,
,  ,
,  , if there were 5 isolates through the course of 2016. Open symbols represent different strains, identified by VNTR analysis. Abbreviations: AMK: amikacin; AZM: aztreonam; CEF: ceftazidime; CIP: ciprofloxacin; GEN: gentamicin; MER: meropenem; PTZ: piperacillin-tazobactam; SXT: co-trimoxazole (no EUCAST breakpoint); TOB: tobramycin. Disc diffusion assay for antibiotic sensitivity testing was performed once at each time point and EUCAST sensitivity cutoffs are indicated by the bars. Biological triplicates of phage assay are represented individually and bars indicate both neat phage (>2 × 102 PFU/mL) and dilute phage (>2 × 105 PFU/mL) cutoffs. (a) Patient H is an example of a patient colonized with MDR Pa, sensitive only to tobramycin and phage at all 5 time points throughout 2016. (b) Patient B represents a patient harboring a Pa strain showing high variability to all antibiotics as well as the phage cocktail. Of note, this patient’s first isolate was resistant to the phage cocktail, while subsequent isolates throughout the year showed susceptibility. (c) Patient D harbored a Pa strain resistant to the phage cocktail at the first two timepoints in the year, but sensitive throughout the rest of 2016. (d) Patient F was one of three from whom two distinct Pa strains were identified. The first two isolates in the year are one strain, while the subsequent two are another. There are distinct antibiograms for the two strains, as well as a distinct pattern of susceptibility/resistance to the phage cocktail.
, if there were 5 isolates through the course of 2016. Open symbols represent different strains, identified by VNTR analysis. Abbreviations: AMK: amikacin; AZM: aztreonam; CEF: ceftazidime; CIP: ciprofloxacin; GEN: gentamicin; MER: meropenem; PTZ: piperacillin-tazobactam; SXT: co-trimoxazole (no EUCAST breakpoint); TOB: tobramycin. Disc diffusion assay for antibiotic sensitivity testing was performed once at each time point and EUCAST sensitivity cutoffs are indicated by the bars. Biological triplicates of phage assay are represented individually and bars indicate both neat phage (>2 × 102 PFU/mL) and dilute phage (>2 × 105 PFU/mL) cutoffs. (a) Patient H is an example of a patient colonized with MDR Pa, sensitive only to tobramycin and phage at all 5 time points throughout 2016. (b) Patient B represents a patient harboring a Pa strain showing high variability to all antibiotics as well as the phage cocktail. Of note, this patient’s first isolate was resistant to the phage cocktail, while subsequent isolates throughout the year showed susceptibility. (c) Patient D harbored a Pa strain resistant to the phage cocktail at the first two timepoints in the year, but sensitive throughout the rest of 2016. (d) Patient F was one of three from whom two distinct Pa strains were identified. The first two isolates in the year are one strain, while the subsequent two are another. There are distinct antibiograms for the two strains, as well as a distinct pattern of susceptibility/resistance to the phage cocktail.
 , with subsequent isolates
, with subsequent isolates  ,
,  ,
,  ,
,  , if there were 5 isolates through the course of 2016. Open symbols represent different strains, identified by VNTR analysis. Abbreviations: AMK: amikacin; AZM: aztreonam; CEF: ceftazidime; CIP: ciprofloxacin; GEN: gentamicin; MER: meropenem; PTZ: piperacillin-tazobactam; SXT: co-trimoxazole (no EUCAST breakpoint); TOB: tobramycin. Disc diffusion assay for antibiotic sensitivity testing was performed once at each time point and EUCAST sensitivity cutoffs are indicated by the bars. Biological triplicates of phage assay are represented individually and bars indicate both neat phage (>2 × 102 PFU/mL) and dilute phage (>2 × 105 PFU/mL) cutoffs. (a) Patient H is an example of a patient colonized with MDR Pa, sensitive only to tobramycin and phage at all 5 time points throughout 2016. (b) Patient B represents a patient harboring a Pa strain showing high variability to all antibiotics as well as the phage cocktail. Of note, this patient’s first isolate was resistant to the phage cocktail, while subsequent isolates throughout the year showed susceptibility. (c) Patient D harbored a Pa strain resistant to the phage cocktail at the first two timepoints in the year, but sensitive throughout the rest of 2016. (d) Patient F was one of three from whom two distinct Pa strains were identified. The first two isolates in the year are one strain, while the subsequent two are another. There are distinct antibiograms for the two strains, as well as a distinct pattern of susceptibility/resistance to the phage cocktail.
, if there were 5 isolates through the course of 2016. Open symbols represent different strains, identified by VNTR analysis. Abbreviations: AMK: amikacin; AZM: aztreonam; CEF: ceftazidime; CIP: ciprofloxacin; GEN: gentamicin; MER: meropenem; PTZ: piperacillin-tazobactam; SXT: co-trimoxazole (no EUCAST breakpoint); TOB: tobramycin. Disc diffusion assay for antibiotic sensitivity testing was performed once at each time point and EUCAST sensitivity cutoffs are indicated by the bars. Biological triplicates of phage assay are represented individually and bars indicate both neat phage (>2 × 102 PFU/mL) and dilute phage (>2 × 105 PFU/mL) cutoffs. (a) Patient H is an example of a patient colonized with MDR Pa, sensitive only to tobramycin and phage at all 5 time points throughout 2016. (b) Patient B represents a patient harboring a Pa strain showing high variability to all antibiotics as well as the phage cocktail. Of note, this patient’s first isolate was resistant to the phage cocktail, while subsequent isolates throughout the year showed susceptibility. (c) Patient D harbored a Pa strain resistant to the phage cocktail at the first two timepoints in the year, but sensitive throughout the rest of 2016. (d) Patient F was one of three from whom two distinct Pa strains were identified. The first two isolates in the year are one strain, while the subsequent two are another. There are distinct antibiograms for the two strains, as well as a distinct pattern of susceptibility/resistance to the phage cocktail.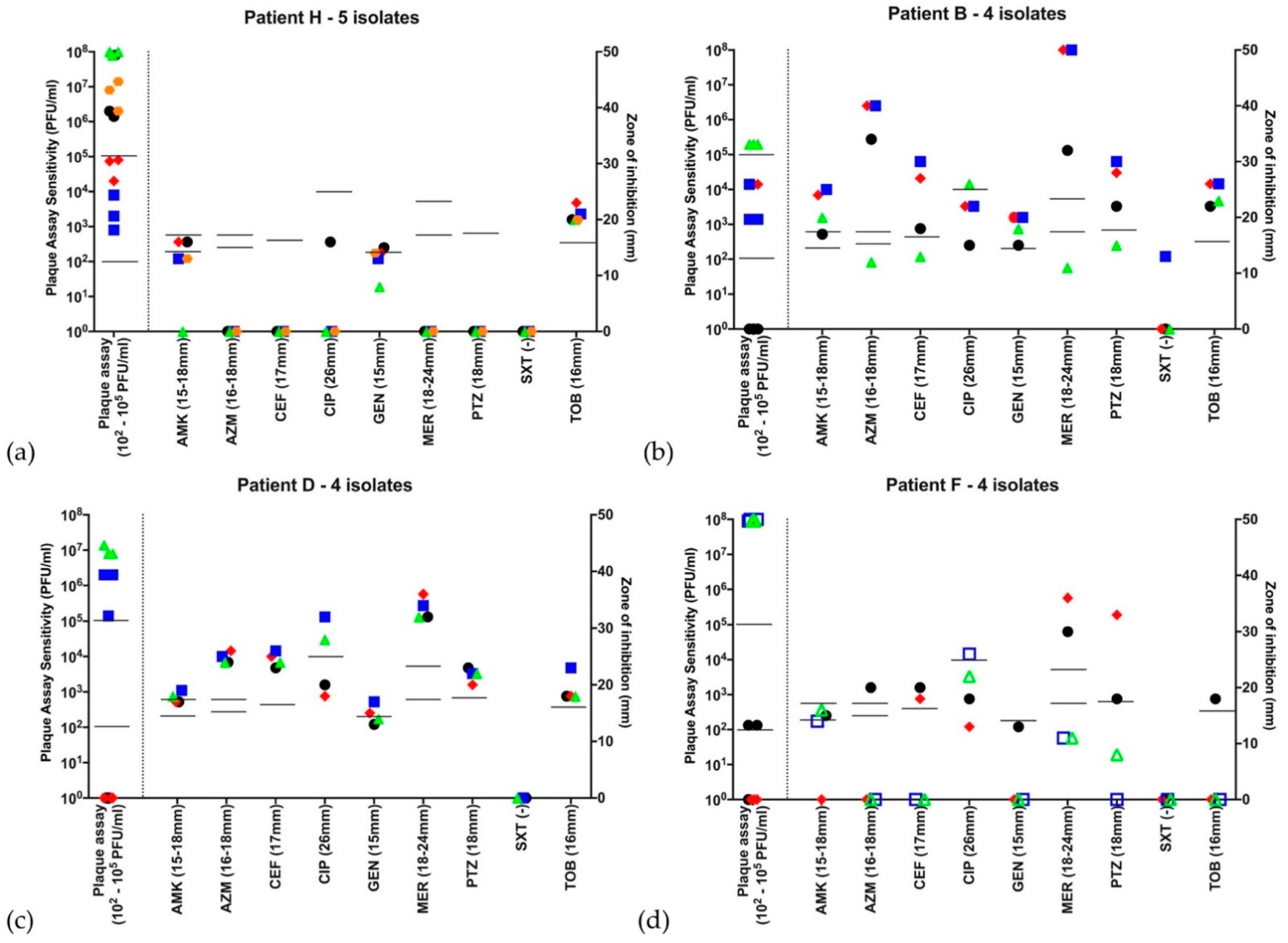
| Patients: | Number | 24 |
|---|---|---|
| Age (median (range)) in years | 31 (14–57) | |
| Male:female ratio | 12:12 | |
| Best FEV1 (% predicted median and range) for 2016 | 62 (23–120) | |
| Number of isolates per patient (median (range)) | 4 (3–6) | |
| Number (%) of subjects with exclusively non-mucoid strains | 3 (12.5%) | |
| Number (%) of subjects with exclusively mucoid strains | 6 (25%) | |
| Number (%) with >1 VNTR type over time period examined | 3 (12.5%) | |
| Subjects (n) with shared strains: Liverpool Epidemic Strain * | 4 | |
| Cluster A * | 2 | |
| Isolates: | Number of isolates (total) | 112 |
| Number with antibiotic and phage sensitivity + VNTR | 102 | |
| Number (%) of non-mucoid/ mucoid isolates | 44 (43%)/ 58 (57%) | |
| Number (%) sensitive to neat phage | 76 (75%) | |
| Number (%) sensitive to dilute phage (10−3 dilution) | 51 (50%) | |
| Number (%) with resistance to ≥1 antibiotic | 88 (86%) | |
| Number (%) meeting definition of MDR ** | 51 (50%) | |
| Number (%) of MDR isolates sensitive to neat phage | 40/51 (78%) | |
| Number (%) of MDR isolates sensitive to dilute phage (10−3 dilution) | 26/51 (51%) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Martin, I.; Kenna, D.T.D.; Morales, S.; Alton, E.W.F.W.; Davies, J.C. Variability in Bacteriophage and Antibiotic Sensitivity in Serial Pseudomonas aeruginosa Isolates from Cystic Fibrosis Airway Cultures over 12 Months. Microorganisms 2021, 9, 660. https://doi.org/10.3390/microorganisms9030660
Martin I, Kenna DTD, Morales S, Alton EWFW, Davies JC. Variability in Bacteriophage and Antibiotic Sensitivity in Serial Pseudomonas aeruginosa Isolates from Cystic Fibrosis Airway Cultures over 12 Months. Microorganisms. 2021; 9(3):660. https://doi.org/10.3390/microorganisms9030660
Chicago/Turabian StyleMartin, Isaac, Dervla T. D. Kenna, Sandra Morales, Eric W. F. W. Alton, and Jane C. Davies. 2021. "Variability in Bacteriophage and Antibiotic Sensitivity in Serial Pseudomonas aeruginosa Isolates from Cystic Fibrosis Airway Cultures over 12 Months" Microorganisms 9, no. 3: 660. https://doi.org/10.3390/microorganisms9030660
APA StyleMartin, I., Kenna, D. T. D., Morales, S., Alton, E. W. F. W., & Davies, J. C. (2021). Variability in Bacteriophage and Antibiotic Sensitivity in Serial Pseudomonas aeruginosa Isolates from Cystic Fibrosis Airway Cultures over 12 Months. Microorganisms, 9(3), 660. https://doi.org/10.3390/microorganisms9030660






