Understanding the Interactions between Staphylococcus aureus and the Raw-Meat-Processing Environment Isolate Klebsiella oxytoca in Dual-Species Biofilms via Discovering an Altered Metabolic Profile
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Media
2.2. Development of Mono- and Dual-Species Biofilms and Quantification
2.3. Field Emission Scanning Electron Microscope (FESEM) Analysis
2.4. Biofilm Collection and Metabolite Extraction
2.5. HILIC UHPLC-Q-TOF MS Analysis
2.6. Multivariate Data Processing and Data Analysis
3. Results
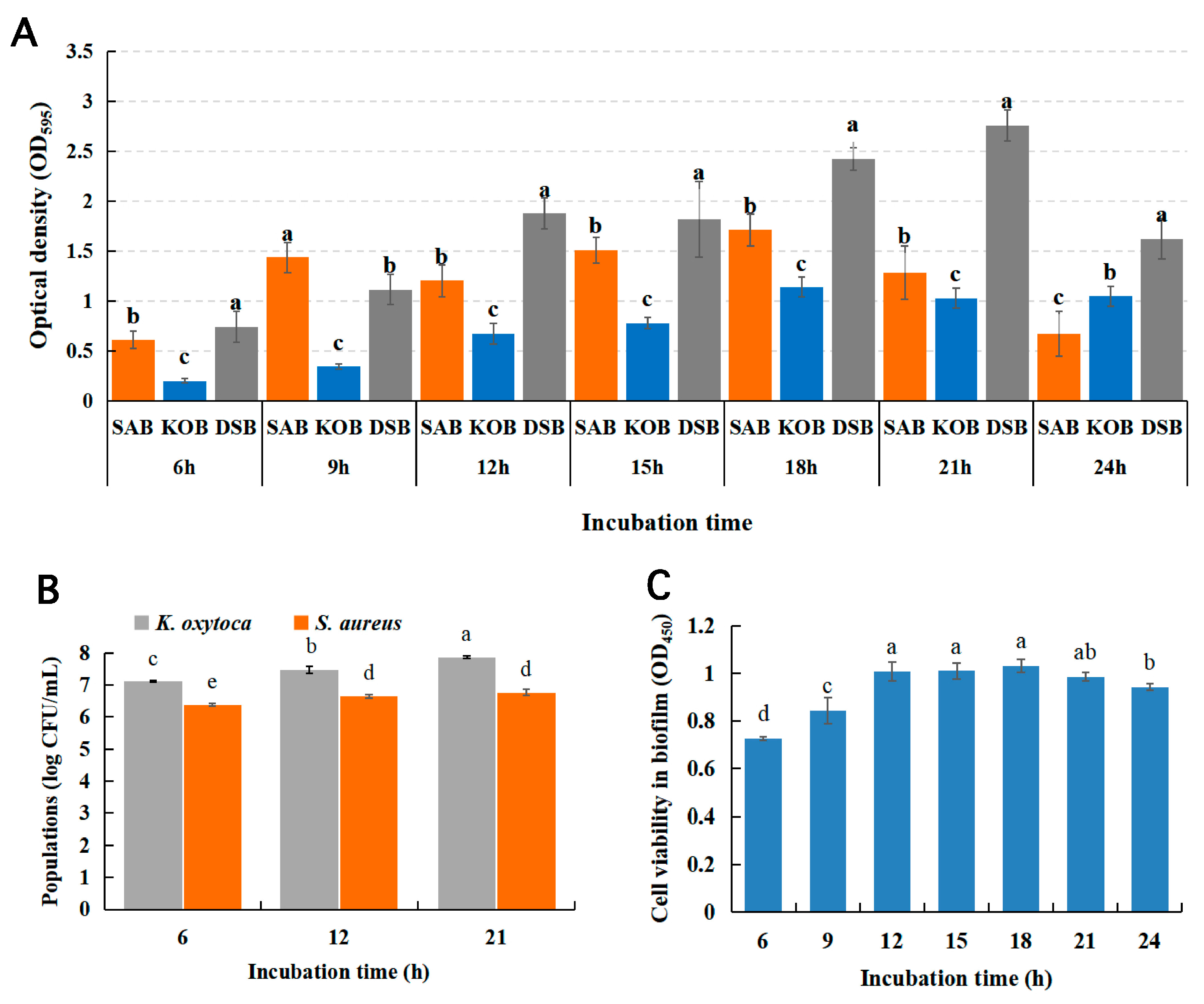
3.1. Characterization of Mono- and Dual-Species Biofilms
3.2. Metabolic Profiling by UHPLC-Q-TOF/MS
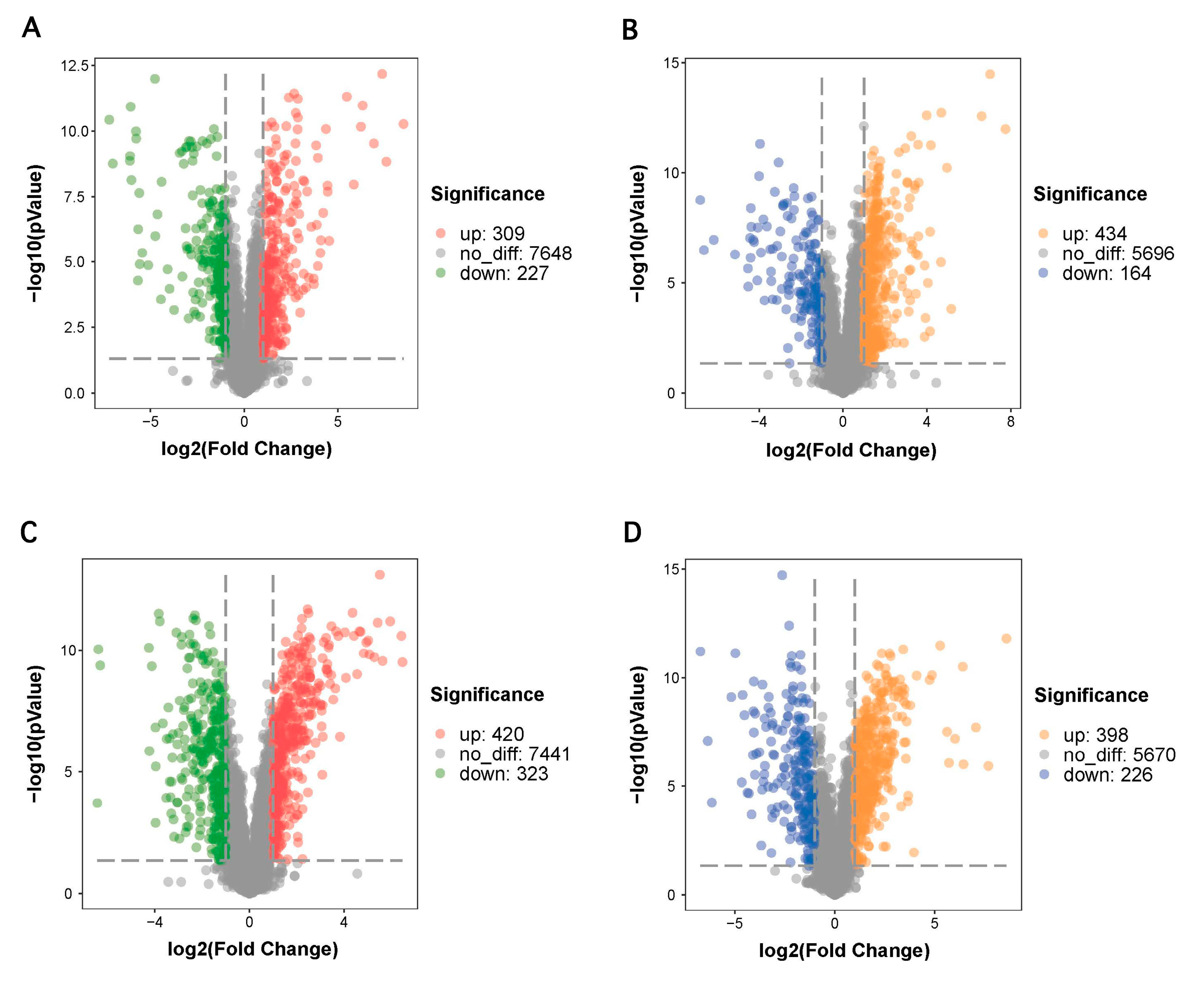
3.3. Screening of Potential Biomarkers
3.4. KEGG Pathway Enrichment Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Simoes, L.C.; Simoes, M.; Vieira, M.J. Biofilm interactions between distinct bacterial genera isolated from drinking water. Appl. Environ. Microbiol. 2007, 73, 6192–6200. [Google Scholar] [CrossRef]
- Kocot, A.M.; Olszewska, M.A. Interaction of Pseudomonas aeruginosa and Staphylococcus aureus with Listeria innocua in dual species biofilms and inactivation following disinfectant treatments. Lwt 2020, 118. [Google Scholar] [CrossRef]
- Kang, J.; Jin, W.; Wang, J.; Sun, Y.; Wu, X.; Liu, L. Antibacterial and anti-biofilm activities of peppermint essential oil against Staphylococcus aureus. LWT 2019, 101, 639–645. [Google Scholar] [CrossRef]
- Fetsch, A.; Contzen, M.; Hartelt, K.; Kleiser, A.; Maassen, S.; Rau, J.; Kraushaar, B.; Layer, F.; Strommenger, B. Staphylococcus aureus food-poisoning outbreak associated with the consumption of ice-cream. Int. J. Food Microbiol. 2014, 187, 1–6. [Google Scholar] [CrossRef]
- Rode, T.M.; Langsrud, S.; Holck, A.; Moretro, T. Different patterns of biofilm formation in Staphylococcus aureus under food-related stress conditions. Int. J. Food Microbiol. 2007, 116, 372–383. [Google Scholar] [CrossRef]
- Wang, H.; Qi, J.; Dong, Y.; Li, Y.; Xu, X.; Zhou, G. Characterization of attachment and biofilm formation by meat-borne Enterobacteriaceae strains associated with spoilage. LWT 2017, 86, 399–407. [Google Scholar] [CrossRef]
- Cai, L.; Wang, H.; Liang, L.; Wang, G.; Xu, X.; Wang, H. Response of Formed-Biofilm of Enterobacter cloacae, Klebsiella oxytoca, and Citrobacter freundii to Chlorite-Based Disinfectants. J. Food Sci. 2018, 83, 1326–1332. [Google Scholar] [CrossRef]
- Gundogan, N.; Citak, S.; Yalcin, E. Virulence Properties of Extended Spectrum β-Lactamase–Producing Klebsiella Species in Meat Samples. J. Food Prot. 2011, 74, 559–564. [Google Scholar] [CrossRef] [PubMed]
- Tondo, E.C.; Lakus, F.R.; Oliveira, F.A.; Brandelli, A. Identification of heat stable protease of Klebsiella oxytoca isolated from raw milk. Lett. Appl. Microbiol. 2004, 38, 146–150. [Google Scholar] [CrossRef]
- Podschun, P.; Ullmann, U. Klebsiella spp. as nosocomial pathogens: Epidemiology, taxonomy, typing methods, and pathogenicity factors. Clin. Microbiol. Rev. 1998, 11, 589–603. [Google Scholar] [CrossRef]
- Govaert, M.; Smet, C.; Walsh, J.L.; Van Impe, J.F.M. Dual-Species Model Biofilm Consisting of Listeria monocytogenes and Salmonella Typhimurium: Development and Inactivation with Cold Atmospheric Plasma (CAP). Front. Microbiol. 2019, 10, 2524. [Google Scholar] [CrossRef]
- Da, W.; Shao, J.; Li, Q.; Shi, G.; Wang, T.; Wu, D.; Wang, C. Extraction of Extracellular Matrix in Static and Dynamic Candida Biofilms Using Cation Exchange Resin and Untargeted Analysis of Matrix Metabolites by Ultra-High-Performance Liquid Chromatography-Tandem Quadrupole Time-of-Flight Mass Spectrometry (UPLC-Q-TOF-MS). Front. Microbiol. 2019, 10, 752. [Google Scholar] [CrossRef]
- Sanders, D.; Borys, K.D.; Kisa, F.; Rakowski, S.A.; Lozano, M.; Filutowicz, M. Multiple Dictyostelid Species Destroy Biofilms of Klebsiella oxytoca and Other Gram Negative Species. Protist 2017, 168, 311–325. [Google Scholar] [CrossRef] [PubMed]
- Burmolle, M.; Ren, D.; Bjarnsholt, T.; Sorensen, S.J. Interactions in multispecies biofilms: Do they actually matter? Trends Microbiol. 2014, 22, 84–91. [Google Scholar] [CrossRef] [PubMed]
- Doulgeraki, A.I.; Di Ciccio, P.; Ianieri, A.; Nychas, G.E. Methicillin-resistant food-related Staphylococcus aureus: A review of current knowledge and biofilm formation for future studies and applications. Res. Microbiol. 2017, 168, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.; Flint, S.H.; Bennett, R.J.; Brooks, J.D.; Morton, R.H. Biofilm growth of individual and dual strains of Klebsiella oxytoca from the dairy industry on ultrafiltration membranes. J. Ind. Microbiol. Biotechnol. 2009, 36, 1491–1497. [Google Scholar] [CrossRef] [PubMed]
- Burmolle, M.; Webb, J.S.; Rao, D.; Hansen, L.H.; Sorensen, S.J.; Kjelleberg, S. Enhanced biofilm formation and increased resistance to antimicrobial agents and bacterial invasion are caused by synergistic interactions in multispecies biofilms. Appl. Environ. Microbiol. 2006, 72, 3916–3923. [Google Scholar] [CrossRef]
- Mashego, M.R.; Rumbold, K.; De Mey, M.; Vandamme, E.; Soetaert, W.; Heijnen, J.J. Microbial metabolomics: Past, present and future methodologies. Biotechnol. Lett. 2007, 29, 1–16. [Google Scholar] [CrossRef]
- Kart, D.; Yabanoglu Ciftci, S.; Nemutlu, E. Altered metabolomic profile of dual-species biofilm: Interactions between Proteus mirabilis and Candida albicans. Microbiol. Res. 2020, 230, 126346. [Google Scholar] [CrossRef]
- Tian, X.; Yu, Q.; Yao, D.; Shao, L.; Liang, Z.; Jia, F.; Li, X.; Hui, T.; Dai, R. New Insights into the Response of Metabolome of Escherichia coli O157:H7 to Ohmic Heating. Front. Microbiol. 2018, 9, 2936. [Google Scholar] [CrossRef]
- Li, H.; Xia, X.; Li, X.; Naren, G.; Fu, Q.; Wang, Y.; Wu, C.; Ding, S.; Zhang, S.; Jiang, H.; et al. Untargeted Metabolomic Profiling of Amphenicol-Resistant Campylobacter jejuni by Ultra-High-Performance Liquid Chromatography–Mass Spectrometry. J. Proteome Res. 2014, 14, 1060–1068. [Google Scholar] [CrossRef]
- Sadiq, F.A.; Flint, S.; Sakandar, H.A.; He, G. Molecular regulation of adhesion and biofilm formation in high and low biofilm producers of Bacillus licheniformis using RNA-Seq. Biofouling 2019, 35, 143–158. [Google Scholar] [CrossRef] [PubMed]
- Velmourougane, K.; Prasanna, R. Influence of l-amino acids on aggregation and biofilm formation in Azotobacter chroococcum and Trichoderma viride. J. Appl. Microbiol. 2017, 123, 977–991. [Google Scholar] [CrossRef]
- Hernandez, S.B.; Cava, F. Environmental roles of microbial amino acid racemases. Environ. Microbiol. 2016, 18, 1673–1685. [Google Scholar] [CrossRef] [PubMed]
- Cava, F.; Lam, H.; de Pedro, M.A.; Waldor, M.K. Emerging knowledge of regulatory roles of D-amino acids in bacteria. Cell Mol. Life Sci. 2011, 68, 817–831. [Google Scholar] [CrossRef]
- Barrientos-Moreno, L.; Molina-Henares, M.A.; Ramos-Gonzalez, M.I.; Espinosa-Urgel, M. Arginine as an environmental and metabolic cue for cyclic diguanylate signalling and biofilm formation in Pseudomonas putida. Sci. Rep. 2020, 10, 13623. [Google Scholar] [CrossRef]
- Vuong, C.; Kidder, J.B.; Jacobson, E.R.; Otto, M.; Proctor, R.A.; Somerville, G.A. Staphylococcus epidermidis polysaccharide intercellular adhesin production significantly increases during tricarboxylic acid cycle stress. J. Bacteriol. 2005, 187, 2967–2973. [Google Scholar] [CrossRef]
- De Backer, S.; Sabirova, J.; De Pauw, I.; De Greve, H.; Hernalsteens, J.P.; Goossens, H.; Malhotra-Kumar, S. Enzymes Catalyzing the TCA- and Urea Cycle Influence the Matrix Composition of Biofilms Formed by Methicillin-Resistant Staphylococcus aureus USA300. Microorganisms 2018, 6, 113. [Google Scholar] [CrossRef] [PubMed]
- Shanks, R.M.; Meehl, M.A.; Brothers, K.M.; Martinez, R.M.; Donegan, N.P.; Graber, M.L.; Cheung, A.L.; O’Toole, G.A. Genetic evidence for an alternative citrate-dependent biofilm formation pathway in Staphylococcus aureus that is dependent on fibronectin binding proteins and the GraRS two-component regulatory system. Infect. Immun. 2008, 76, 2469–2477. [Google Scholar] [CrossRef]
- Liu, C.; Sun, D.; Zhu, J.; Liu, W. Two-Component Signal Transduction Systems: A Major Strategy for Connecting Input Stimuli to Biofilm Formation. Front. Microbiol. 2018, 9, 3279. [Google Scholar] [CrossRef]
- Wu, S.; Lin, K.; Liu, Y.; Zhang, H.; Lei, L. Two-component signaling pathways modulate drug resistance of Staphylococcus aureus (Review). Biomed. Rep. 2020, 13, 5. [Google Scholar] [CrossRef]
- Nagasawa, R.; Sato, T.; Senpuku, H. Raffinose induces biofilm formation by Streptococcus mutans in low concentrations of sucrose by increasing production of extracellular DNA and fructan. Appl. Environ. Microbiol. 2017, 83. [Google Scholar] [CrossRef]
- Nuraini, P.; Pradopo, S.; Pronorahardjo, A.S. Sucrose and Xylitol-Induced Streptococcus mutans Biofilm Adherence. Pesqui. Bras. Em Odontopediatria e Clínica Integr. 2020, 20. [Google Scholar] [CrossRef]




| Mode | No | Adduct | Rt(s) | m/z | Metabolite | VIP | Fold Change | p-Value |
|---|---|---|---|---|---|---|---|---|
| ESI (+) | 1 | (M + H) + | 475.2005 | 166.0848377 | L-phenylalanine | 17.7719 | 7.253863106 | 0.000 |
| 2 | (M + H)+ | 770.426 | 148.0585192 | L-glutamate | 2.62697 | 6.590693393 | 0.000 | |
| 3 | (M + H)+ | 651.8765 | 90.05302762 | L-alanine | 1.73871 | 3.383882682 | 0.011 | |
| 4 | (M + H)+ | 510.724 | 132.1001379 | L-isoleucine | 6.32109 | 3.021382002 | 0.000 | |
| 5 | (M + H)+ | 956.207 | 175.1174382 | L-arginine | 1.06786 | 1.463831102 | 0.036 | |
| 6 | (M + H)+ | 565.397 | 118.084813 | L-valine | 3.23266 | 1.41510846 | 0.003 | |
| 7 | (M + NH4)+ | 861.9085 | 522.2015198 | Raffinose | 1.47743 | 0.875635638 | 0.017 | |
| 8 | (M + H-H2O)+ | 226.93 | 129.0638672 | L-glutamine | 1.84291 | 0.735582718 | 0.001 | |
| 9 | (M + H)+ | 504.573 | 118.0849979 | Betaine | 7.42027 | 0.693008883 | 0.019 | |
| 10 | (M + H)+ | 906.151 | 184.0714508 | Phosphorylcholine | 1.51653 | 0.214757887 | 0.000 | |
| ESI (–) | 1 | (M-H)- | 470.852 | 164.0712635 | L-phenylalanine | 12.9064 | 11.88920095 | 0.000 |
| 2 | (M-H)- | 745.958 | 129.0184177 | Citraconic acid | 1.24581 | 0.167383867 | 0.000 | |
| 3 | (M-H2O-H)- | 510.2385 | 152.9949551 | Glycerol 3-phosphate | 1.22255 | 3.659826379 | 0.000 | |
| 4 | (M-H)- | 900.126 | 191.0192075 | Citrate | 1.30013 | 4.143342579 | 0.000 | |
| 5 | (M-H)- | 428.5495 | 89.02377085 | DL-lactate | 2.68168 | 15.2069271 | 0.000 | |
| 6 | (M-H)- | 505.751 | 130.0866943 | L-leucine | 6.6272 | 1.700068517 | 0.002 | |
| 7 | (M-H)- | 744.368 | 179.0556474 | Myo-inositol | 1.14716 | 1.141321917 | 0.003 | |
| 8 | (M-H)- | 830.001 | 154.0615257 | L-histidine | 1.82022 | 3.032862317 | 0.008 |
| Mode | No | Adduct | Rt(s) | m/z | Metabolite | VIP | Fold Change | p-Value |
|---|---|---|---|---|---|---|---|---|
| ESI (+) | 1 | (M + H)+ | 956.207 | 175.1174382 | L-arginine | 20.8719 | 86.28151666 | 0.000 |
| 2 | (M + H)+ | 651.4535 | 132.0640863 | Hydroxyproline | 7.84314 | 0.136447311 | 0.000 | |
| 3 | (M + H)+ | 584.999 | 116.0687626 | D-proline | 9.17181 | 8.063013806 | 0.000 | |
| 4 | (M+NH4)+ | 861.9085 | 522.2015198 | Raffinose | 4.22294 | 0.358515007 | 0.000 | |
| 5 | (M + H)+ | 580.177 | 131.1162654 | N-acetylputrescine | 16.9024 | 0.167206532 | 0.000 | |
| 6 | (M + H)+ | 651.8765 | 90.05302762 | L-alanine | 3.53333 | 7.029141017 | 0.000 | |
| 7 | (M + H)+ | 684.3185 | 76.03758772 | Glycine | 2.0612 | 8.323423876 | 0.000 | |
| 8 | (M + H)+ | 783.3 | 134.0430189 | L-aspartate | 1.29637 | 3.978271643 | 0.000 | |
| 9 | (M + H)+ | 669.03 | 146.0908832 | 4-Guanidinobutyric acid | 1.05007 | 0.786705284 | 0.010 | |
| 10 | (M + NH4)+ | 738.9845 | 360.1485731 | Sucrose | 1.56376 | 1.879752696 | 0.010 | |
| 11 | (M + H-H2O)+ | 226.93 | 129.0638672 | L-glutamine | 1.13001 | 1.155452429 | 0.034 | |
| ESI (–) | 1 | (M-H)- | 982.1585 | 173.1038206 | L-arginine | 7.29498 | 84.61874522 | 0.000 |
| 2 | (M-H)- | 645.4265 | 130.0499991 | Hydroxyproline | 1.44552 | 0.235656929 | 0.000 | |
| 3 | (M-H)- | 646.5645 | 88.03968055 | Sarcosine | 2.23067 | 11.0313482 | 0.000 | |
| 4 | (M-H)- | 483.812 | 130.086757 | L-leucine | 4.44469 | 0.092413693 | 0.000 | |
| 5 | (M-H)- | 428.5495 | 89.02377085 | DL-lactate | 5.46247 | 64.61340268 | 0.000 | |
| 6 | (M-H)- | 736.06 | 117.0184715 | Succinate | 5.20026 | 52.41791755 | 0.000 | |
| 7 | (M-H)- | 730.801 | 341.1082907 | Sucrose | 2.42076 | 7.285447139 | 0.000 | |
| 8 | (M-H)- | 830.001 | 154.0615257 | L-histidine | 2.048 | 3.51164159 | 0.000 | |
| 9 | (M-H)- | 959.703 | 191.0190107 | Citrate | 1.14805 | 15.54534738 | 0.011 | |
| 10 | (M-H)- | 555.6405 | 116.0709687 | L-valine | 1.63014 | 0.868488008 | 0.031 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, X.; Hu, Y.; Tian, S.; Han, B. Understanding the Interactions between Staphylococcus aureus and the Raw-Meat-Processing Environment Isolate Klebsiella oxytoca in Dual-Species Biofilms via Discovering an Altered Metabolic Profile. Microorganisms 2021, 9, 672. https://doi.org/10.3390/microorganisms9040672
Chen X, Hu Y, Tian S, Han B. Understanding the Interactions between Staphylococcus aureus and the Raw-Meat-Processing Environment Isolate Klebsiella oxytoca in Dual-Species Biofilms via Discovering an Altered Metabolic Profile. Microorganisms. 2021; 9(4):672. https://doi.org/10.3390/microorganisms9040672
Chicago/Turabian StyleChen, Xiaoxue, Yunan Hu, Simin Tian, and Beizhong Han. 2021. "Understanding the Interactions between Staphylococcus aureus and the Raw-Meat-Processing Environment Isolate Klebsiella oxytoca in Dual-Species Biofilms via Discovering an Altered Metabolic Profile" Microorganisms 9, no. 4: 672. https://doi.org/10.3390/microorganisms9040672
APA StyleChen, X., Hu, Y., Tian, S., & Han, B. (2021). Understanding the Interactions between Staphylococcus aureus and the Raw-Meat-Processing Environment Isolate Klebsiella oxytoca in Dual-Species Biofilms via Discovering an Altered Metabolic Profile. Microorganisms, 9(4), 672. https://doi.org/10.3390/microorganisms9040672





