Diagnostic Performance of a Magnetic Field-Enhanced Agglutination Readout in Detecting Either Viral Genomes or Host Antibodies in Arbovirus Infection
Abstract
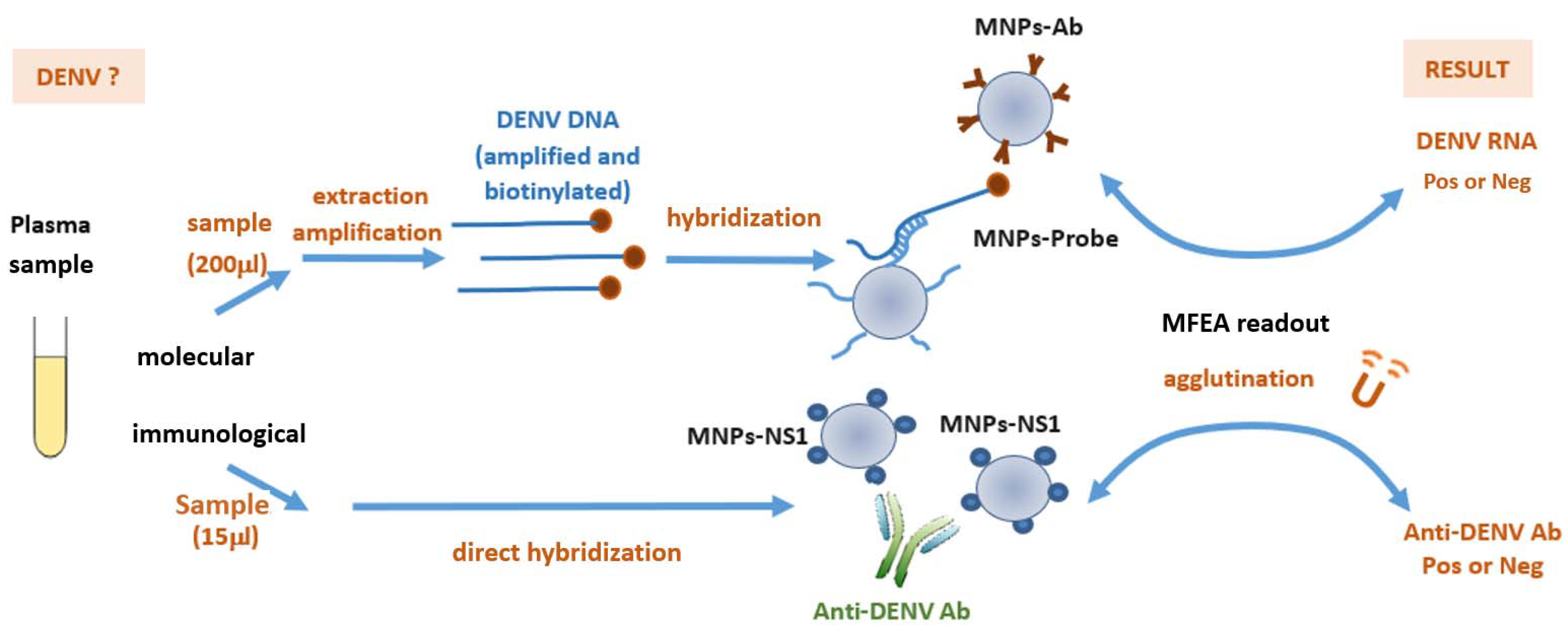
:1. Introduction
2. Materials and Methods
2.1. Biological Samples
2.2. Viral Nucleic Acid Extraction and Amplification
2.3. Grafting of Tetrathiolated DENV Probes onto Magnetic Nanoparticles
2.4. Grafting of DENV NS1 Antigens onto Magnetic Nanoparticles
2.5. Molecular MFEA Readout
2.6. Immunological MFEA Readout
2.7. Statistical Analysis
3. Results
3.1. Sample Characteristics
3.2. Diagnostic Performance of the Molecular MFEA Readout
- Negative plasma from donors (n = 32) and positive plasma samples from patients (n = 43) were assayed. The turbidity signal is expressed as the difference of optical density at 650 nm (ΔOD650nm) measured before and after the three magnetization cycles. The limit of detection (LOD) is taken as the mean value of blank samples plus three standard deviations. Individual points of the scatterplot represent the ratio of turbidity signal/LOD calculated for one sample by the molecular MFEA readout. Data are expressed as median ratios with interquartile ranges.
| Sample Type | Samples, n | Samples Correctly Detected, n | Diagnostic Sensitivity * % (95% CI) | Diagnostic Specificity † % (95% CI) | Accuracy ‡ % |
|---|---|---|---|---|---|
| DENV | 43 | 38 | 88.37 (78.79–97.95) | / | 92 |
| Healthy | 32 | 31 | / | 96.87 (90.84–100.00) |
3.3. Diagnostic Performance of the Immunological MFEA Readout

| Sample Type | Samples, n | Samples Correctly Detected, n | Diagnostic Sensitivity * % (95% CI) | Diagnostic Specificity † % (95% CI) | Accuracy ‡ % |
|---|---|---|---|---|---|
| DENV | 64 | 58 | 90.62 (83.50–97.76) | / | 93.20 |
| Healthy | 39 | 38 | / | 97.44 (92.48–100.00) |
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Musso, D.; Rodriguez-Morales, A.J.; Levi, J.E.; Cao-Lormeau, V.M.; Gubler, D.J. Unexpected outbreaks of arbovirus infections: Lessons learned from the Pacific and tropical America. Lancet Infect. Dis. 2018, 18, e355–e361. [Google Scholar] [CrossRef]
- Weaver, S.C.; Charlier, C.; Vasilakis, N.; Lecuit, M. Zika, Chikungunya, and Other Emerging Vector-Borne Viral Diseases. Annu. Rev. Med. 2018, 69, 395–408. [Google Scholar] [CrossRef] [PubMed]
- Amato, L.; Dente, M.G.; Calistri, P.; Declich, S.; On Behalf Of The MediLabSecure Working Group. Integrated Early Warning Surveillance: Achilles’ Heel of One Health? Microorganisms 2020, 8, 84. [Google Scholar] [CrossRef]
- Guzman, M.G.; Gubler, D.J.; Izquierdo, A.; Martinez, E.; Halstead, S.B. Dengue infection. Nat. Rev. Dis. Primers 2016, 2, 16055. [Google Scholar] [CrossRef]
- Katzelnick, L.C.; Coloma, J.; Harris, E. Dengue: Knowledge gaps, unmet needs, and research priorities. Lancet Infect. Dis 2017, 17, e88–e100. [Google Scholar] [CrossRef]
- Jourdain, F.; Samy, A.M.; Hamidi, A.; Bouattour, A.; Alten, B.; Faraj, C.; Roiz, D.; Petric, D.; Perez-Ramirez, E.; Velo, E.; et al. Towards harmonisation of entomological surveillance in the Mediterranean area. PLoS Negl. Trop. Dis. 2019, 13, e0007314. [Google Scholar] [CrossRef] [PubMed]
- Peeling, R.W.; Artsob, H.; Pelegrino, J.L.; Buchy, P.; Cardosa, M.J.; Devi, S.; Enria, D.A.; Farrar, J.; Gubler, D.J.; Guzman, M.G.; et al. Evaluation of diagnostic tests: Dengue. Nat. Rev. Microbiol. 2010, 8, S30–S38. [Google Scholar] [CrossRef]
- Chen, H.; Liu, K.; Li, Z.; Wang, P. Point of care testing for infectious diseases. Clin. Chim. Acta 2019, 493, 138–147. [Google Scholar] [CrossRef]
- Goncalves, A.; Peeling, R.W.; Chu, M.C.; Gubler, D.J.; de Silva, A.M.; Harris, E.; Murtagh, M.; Chua, A.; Rodriguez, W.; Kelly, C.; et al. Innovative and New Approaches to Laboratory Diagnosis of Zika and Dengue: A Meeting Report. J. Infect. Dis. 2018, 217, 1060–1068. [Google Scholar] [CrossRef] [PubMed]
- Guzman, M.G.; Halstead, S.B.; Artsob, H.; Buchy, P.; Farrar, J.; Gubler, D.J.; Hunsperger, E.; Kroeger, A.; Margolis, H.S.; Martinez, E.; et al. Dengue: A continuing global threat. Nat. Rev. Microbiol. 2010, 8, S7–S16. [Google Scholar] [CrossRef]
- Leon, F.; Meyer, A.; Reynier, R.; Blanc, E.; Bruyere-Ostells, L.; Bres, J.C.; Simonin, Y.; Salinas, S.; Gallian, P.; Leparc-Goffart, I.; et al. An Innovative Multiplexed and Flexible Molecular Approach for the Differential Detection of Arboviruses. J. Mol. Diagn. 2019, 21, 81–88. [Google Scholar] [CrossRef]
- Pinchon, E.; Leon, F.; Temurok, N.; Morvan, F.; Vasseur, J.J.; Clot, M.; Foulongne, V.; Cantaloube, J.F.; Vande Perre, P.; Daynès, A.; et al. Rapid and specific DNA detection by magnetic-field enhanced agglutination assay. Talanta 2020, 219, 121344. [Google Scholar] [CrossRef]
- Musso, D.; Despres, P. Serological Diagnosis of Flavivirus-Associated Human Infections. Diagnostics 2020, 10, 302. [Google Scholar] [CrossRef] [PubMed]
- Scaramozzino, N.; Crance, J.M.; Jouan, A.; DeBriel, D.A.; Stoll, F.; Garin, D. Comparison of flavivirus universal primer pairs and development of a rapid, highly sensitive heminested reverse transcription-PCR assay for detection of flaviviruses targeted to a conserved region of the NS5 gene sequences. J. Clin. Microbiol. 2001, 39, 1922–1927. [Google Scholar] [CrossRef] [PubMed]
- Newcombe, R. Two-sided confidence intervals for the single proportion: Comparison of seven methods. Stat. Med. 1998, 17, 857–872. [Google Scholar] [CrossRef]
- Armbruster, D.A.; Pry, T. Limit of blank, limit of detection and limit of quantitation. Clin. Biochem. Rev. 2008, 29 (Suppl. 1), S49–S52. [Google Scholar]
- Barzon, L. Ongoing and emerging arbovirus threats in Europe. J. Clin. Virol. 2018, 107, 38–47. [Google Scholar] [CrossRef]
- Petersen, E.; Petrosillo, N.; Koopmans, M.; Beeching, N.; Di Caro, A.; Gkrania-Klotsas, E.; Kantele, A.; Kohlmann, R.; Lim, P.-L.; Markotic, A.; et al. Emerging infections-an increasingly important topic: Review by the Emerging Infections Task Force. Clin. Microbiol. Infect. 2018, 24, 369–375. [Google Scholar] [CrossRef]
- Bournazos, S.; Gupta, A.; Ravetch, J.V. The role of IgG Fc receptors in antibody-dependent enhancement. Nat. Rev. Immunol. 2020, 20, 633–643. [Google Scholar] [CrossRef]
- Katzelnick, L.C.; Narvaez, C.; Arguello, S.; Lopez Mercado, B.; Collado, D.; Ampie, O.; Elizondo, D.; Miranda, T.; Bustos Carillo, F.; Mercado, J.C.; et al. Zika virus infection enhances future risk of severe dengue disease. Science 2020, 369, 1123–1128. [Google Scholar] [CrossRef]
- Begum, F.; Das, S.; Mukherjee, D.; Ray, U. Hijacking the Host Immune Cells by Dengue Virus: Molecular Interplay of Receptors and Dengue Virus Envelope. Microorganisms 2019, 7, 323. [Google Scholar] [CrossRef] [PubMed]
- Morjaria, S.; Chapin, K.C. Who to Test, When, and for What: Why Diagnostic Stewardship in Infectious Diseases Matters. J. Mol. Diagn. 2020, 22, 1109–1113. [Google Scholar] [CrossRef] [PubMed]
- Peeling, R.W.; Smith, P.G.; Bossuyt, P.M. A guide for diagnostic evaluations. Nat. Rev. Microbiol. 2010, 8, S2–S6. [Google Scholar] [CrossRef] [PubMed]
- Suea-Ngam, A.; Bezinge, L.; Mateescu, B.; Howes, P.D.; deMello, A.J.; Richards, D.A. Enzyme-Assisted Nucleic Acid Detection for Infectious Disease Diagnostics: Moving toward the Point-of-Care. ACS Sens 2020, 5, 2701–2723. [Google Scholar] [CrossRef] [PubMed]
- Ranzoni, A.; Schleipen, J.J.; van Ijzendoorn, L.J.; Prins, M.W. Frequency-selective rotation of two-particle nanoactuators for rapid and sensitive detection of biomolecules. Nano Lett. 2011, 11, 2017–2022. [Google Scholar] [CrossRef]
- Gloag, L.; Mehdipour, M.; Chen, D.; Tilley, R.D.; Gooding, J.J. Advances in the Application of Magnetic Nanoparticles for Sensing. Adv. Mater. 2019, 31, e1904385. [Google Scholar] [CrossRef]
- Schrittwieser, S.; Pelaz, B.; Parak, W.J.; Lentijo-Mozo, S.; Soulantica, K.; Dieckhoff, J.; Ludwig, F.; Guenther, A.; Tschope, A.; Schotter, J. Homogeneous Biosensing Based on Magnetic Particle Labels. Sensors 2016, 16, 828. [Google Scholar] [CrossRef] [PubMed]
- Alejo-Cancho, I.; Navero-Castillejos, J.; Peiro-Mestres, A.; Albarracin, R.; Barrachina, J.; Navarro, A.; Gonzalo, V.; Pastor, V.; Munoz, J.; Martinez, M.J. Evaluation of a novel microfluidic immuno-magnetic agglutination assay method for detection of dengue virus NS1 antigen. PLoS Negl. Trop. Dis. 2020, 14, e0008082. [Google Scholar] [CrossRef]
- Mezger, A.; Fock, J.; Antunes, P.; Osterberg, F.W.; Boisen, A.; Nilsson, M.; Hansen, M.F.; Ahlford, A.; Donolato, M. Scalable DNA-Based Magnetic Nanoparticle Agglutination Assay for Bacterial Detection in Patient Samples. ACS Nano 2015, 9, 7374–7382. [Google Scholar] [CrossRef]
- Moerland, C.P.; van, I.L.J.; Prins, M.W.J. Rotating magnetic particles for lab-on-chip applications—A comprehensive review. Lab Chip 2019, 19, 919–933. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Leon, F.; Pinchon, E.; Temurok, N.; Morvan, F.; Vasseur, J.-J.; Clot, M.; Foulongne, V.; Cantaloube, J.-F.; Vande Perre, P.; Molès, J.-P.; et al. Diagnostic Performance of a Magnetic Field-Enhanced Agglutination Readout in Detecting Either Viral Genomes or Host Antibodies in Arbovirus Infection. Microorganisms 2021, 9, 674. https://doi.org/10.3390/microorganisms9040674
Leon F, Pinchon E, Temurok N, Morvan F, Vasseur J-J, Clot M, Foulongne V, Cantaloube J-F, Vande Perre P, Molès J-P, et al. Diagnostic Performance of a Magnetic Field-Enhanced Agglutination Readout in Detecting Either Viral Genomes or Host Antibodies in Arbovirus Infection. Microorganisms. 2021; 9(4):674. https://doi.org/10.3390/microorganisms9040674
Chicago/Turabian StyleLeon, Fanny, Elena Pinchon, Nevzat Temurok, François Morvan, Jean-Jacques Vasseur, Martine Clot, Vincent Foulongne, Jean-François Cantaloube, Philippe Vande Perre, Jean-Pierre Molès, and et al. 2021. "Diagnostic Performance of a Magnetic Field-Enhanced Agglutination Readout in Detecting Either Viral Genomes or Host Antibodies in Arbovirus Infection" Microorganisms 9, no. 4: 674. https://doi.org/10.3390/microorganisms9040674






