A Study of 3CLpros as Promising Targets against SARS-CoV and SARS-CoV-2
Abstract
:1. Introduction
2. Materials and Methods
2.1. Protein Expression and Purification of SARS-CoV 3CLpros
2.2. FRET Protease Assays with SARS-CoV 3CLpros
2.3. FRET Protease Assays with SARS-CoV 3CLpros in the Presence of Triton X-100
2.4. Ligand Preparation, Target Preparation and Induced-Fit Docking
3. Results & Discussion
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Zhu, N.; Zhang, D.; Wang, W.; Li, X.; Yang, B.; Song, J.; Zhao, X.; Huang, B.; Shi, W.; Lu, R.; et al. China Novel Coronavirus Investigating and Research Team. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020, 382, 727–733. [Google Scholar] [CrossRef]
- Olsen, S.J.; Chen, M.Y.; Liu, Y.L.; Witschi, M.; Ardoin, A.; Calba, C.; Mathieu, P.; Masserey, V.; Maraglino, F.; Marro, S.; et al. Early introduction of severe acute respiratory syndrome coronavirus 2 into Europe. Emerg Infect. Dis. 2020, 26, 1567. [Google Scholar] [CrossRef] [PubMed]
- Chen, N.; Zhou, M.; Dong, X.; Qu, J.; Gong, F.; Han, Y.; Qiu, Y.; Wang, J.; Liu, Y.; Wei, Y.; et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: A descriptive study. Lancet 2020, 395, 507–513. [Google Scholar] [CrossRef] [Green Version]
- Team TNCPERE. The epidemiological characteristics of an outbreak of 2019 novel coronavirus diseases (COVID-19) in China. China CDC Wkly. 2020, 41, 145. [Google Scholar]
- Wu, P.; Hao, X.; Lau, E.H.; Wong, J.Y.; Leung, K.S.; Wu, J.T.; Cowling, B.J.; Leung, G.M. Real-time tentative assessment of the epidemiological characteristics of novel coronavirus infections in Wuhan, China, as at 22 January 2020. Eurosurveillance 2020, 25, 2000044. [Google Scholar] [CrossRef]
- Biggerstaff, M.; Cauchemez, S.; Reed, C.; Gambhir, M.; Finelli, L. Estimates of the reproduction number for seasonal, pandemic, and zoonotic influenza: A systematic review of the literature. BMC Infect. Dis. 2014, 14, 480. [Google Scholar] [CrossRef] [Green Version]
- Zhao, S.; Lin, Q.; Ran, J.; Musa, S.S.; Yang, G.; Wang, W.; Lou, Y.; Gao, D.; Yang, L.; He, D.; et al. Preliminary estimation of the basic reproduction number of novel coronavirus (2019-nCoV) in China, from 2019 to 2020: A data-driven analysis in the early phase of the outbreak. Int. J. Infect. Dis. 2020, 92, 214–217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jin, Z.; Smith, L.K.; Rajwanshi, V.K.; Kim, B.; Deval, J. The ambiguous base-pairing and high substrate efficiency of T-705 (Favipiravir) Ribofuranosyl 5’-triphosphate towards influenza a virus polymerase. PLoS ONE 2013, 8, e68347. [Google Scholar] [CrossRef]
- Magee, W.C.; Hostetler, K.Y.; Evans, D.H. Mechanism of inhibition of vaccinia virus DNA polymerase by cidofovir diphosphate. Antimicrob. Agents Chemother. 2005, 49, 3153–3162. [Google Scholar] [CrossRef] [Green Version]
- Cruciani, M.; Martí-Carvajal, A.J.; Mengoli, C.; Serpelloni, G.; Bovo, C.; Moyle, G. Abacavir versus other nucleoside reverse transcriptase inhibitor (NRTI) backbone therapies for treatment of HIV infection. Cochrane Database Syst. Rev. 2018, 2018, CD009390. [Google Scholar] [CrossRef] [Green Version]
- Sham, H.L.; Kempf, D.J.; Molla, A.; Marsh, K.C.; Kumar, G.N.; Chen, C.M.; Kati, W.; Stewart, K.; Lal, R.; Hsu, A.; et al. ABT-378, a highly potent inhibitor of the human immunodeficiency virus protease. Antimicrob. Agents Chemother. 1998, 42, 3218–3224. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Agbowuro, A.A.; Huston, W.M.; Gamble, A.B.; Tyndall, J.D.A. Proteases and protease inhibitors in infectious diseases. Med. Res. Rev. 2018, 38, 1295–1331. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Lin, D.; Sun, X.; Curth, U.; Drosten, C.; Sauerhering, L.; Becker, S.; Rox, K.; Hilgenfeld, R. Crystal structure of SARSCoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science 2020, 20, 409–412. [Google Scholar] [CrossRef] [Green Version]
- Su, H.X.; Yao, S.; Zhao, W.F.; Li, M.J.; Liu, J.; Shang, W.J.; Xie, H.; Ke, C.Q.; Hu, H.C.; Gao, M.N.; et al. Anti-SARS-CoV-2 activities in vitro of Shuanghuanglian preparations and bioactive ingredients. Acta Pharmacol. Sin. 2020, 41, 1167–1177. [Google Scholar] [CrossRef]
- Fu, L.; Ye, F.; Feng, Y.; Yu, F.; Wang, Q.; Wu, Y.; Zhao, C.; Sun, H.; Huang, B.; Niu, P.; et al. Both Boceprevir and GC376 efficaciously inhibit SARS-CoV-2 by targeting its main protease. Nat. Commun. 2020, 11, 4417. [Google Scholar] [CrossRef] [PubMed]
- Corsello, S.M.; Bittker, J.A.; Liu, Z.; Gould, J.; McCarren, P.; Hirschman, J.E.; Johnston, S.E.; Vrcic, A.; Wong, B.; Khan, M.; et al. The Drug Repurposing Hub: A next-generation drug library and information resource. Nat. Med. 2017, 23, 405–408. [Google Scholar] [CrossRef] [Green Version]
- Jo, S.; Kim, S.; Kim, D.Y.; Kim, M.; Shin, D.H. Flavonoids with inhibitory activity against SARS-CoV-2 3CLpro. J. Enzym. Inhib. Med. Chem. 2020, 35, 1539–1544. [Google Scholar] [CrossRef] [PubMed]
- Kuo, C.-J.; Chi, Y.-H.; Hsu, J.T.; Liang, P.-H. Characterization of SARS main protease and inhibitor assay using a fluorogenic substrate. Biochem. Biophys. Res. Commun. 2004, 318, 862–867. [Google Scholar] [CrossRef]
- Wu, A.; Wang, Y.I.; Zeng, C.; Huang, X.; Xu, S.; Su, C.; Wang, M.; Chen, Y.; Guo, D. Prediction and biochemical analysis of putative cleavage sites of the 3C-like protease of Middle East respiratory syndrome coronavirus. Virus Res. 2015, 208, 56–65. [Google Scholar] [CrossRef]
- Sherman, W.; Day, T.; Jacobson, M.P.; Friesner, R.A.; Farid, R. Novel Procedure for Modeling Ligand/Receptor Induced Fit Effects. J. Med. Chem. 2006, 49, 534–553. [Google Scholar] [CrossRef]
- Jacobson, M.P.; Pincus, D.L.; Rapp, C.S.; Day, T.J.; Honig, B.; Shaw, D.E.; Friesner, R.A. A hierarchical approach to all-atom protein loop prediction. Proteins 2004, 55, 351–367. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Friesner, R.A.; Murphy, R.B.; Repasky, M.P.; Frye, L.L.; Greenwood, J.R.; Halgren, T.A.; Sanschagrin, P.C.; Mainz, D.T. Extra precision glide: Docking and scoring incorporating a model of hydrophobic enclosure for protein-ligand complexes. J. Med. Chem. 2006, 49, 6177–6196. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, D.; Kumari, K.; Bahadur, I.; Singh, P. Promising Acyclovir and its derivatives to inhibit the protease of SARS-CoV-2: Molecular Docking and Molecular Dynamics simulations. Res. Sq. 2020. [Google Scholar] [CrossRef]
- Jockusch, S.; Tao, C.; Li, X.; Anderson, T.K.; Chien, M.; Kumar, S.; Russo, J.J.; Kirchdoerfer, R.N.; Ju, J. A library of nucleotide analogues terminate RNA synthesis catalyzed by polymerases of coronaviruses that cause SARS and COVID-19. Antivir. Res. 2020, 180, 104857. [Google Scholar] [CrossRef]
- Gurung, A.B.; Ali, M.A.; Lee, J.; Farah, M.A.; Al-Anazi, K.M. Unravelling lead antiviral phytochemicals for the inhibition of SARS-CoV-2 Mpro enzyme through in silico approach. Life Sci. 2020, 255, 117831. [Google Scholar] [CrossRef]
- Shannon, A.; Selisko, B.; Huchting, J.; Touret, F.; Piorkowski, G.; Fattorini, V.; Ferron, F.; Decroly, E.; Meier, C.; Coutard, B.; et al. Rapid incorporation of Favipiravir by the fast and permissive viral RNA polymerase complex results in SARS-CoV-2 lethal mutagenesis. Nat. Commun. 2020, 11, 4682. [Google Scholar] [CrossRef] [PubMed]
- Beck, B.R.; Shin, B.; Choi, Y.; Park, S.; Kang, K. Predicting commercially available antiviral drugs that may act on the novel coronavirus (SARS-CoV-2) through a drug-target interaction deep learning model. Comput. Struct. Biotechnol. J. 2020, 18, 784–790. [Google Scholar] [CrossRef]
- Pruijssers, A.J.; George, A.S.; Schäfer, A.; Leist, S.R.; Gralinksi, L.E.; Dinnon, K.H. Remdesivir potently inhibits SARS-CoV-2 in human lung cells and chimeric SARS-CoV expressing the SARS-CoV-2 RNA polymerase in mice. bioRxiv 2020. [Google Scholar] [CrossRef]
- Jácome, R.; Campillo-Balderas, J.A.; de León, S.P.; Becerra, A.; Lazcano, A. Sofosbuvir as a potential alternative to treat the SARS-CoV-2 epidemic. Sci. Rep. 2020, 10, 9294. [Google Scholar] [CrossRef]
- Indu, P.; Rameshkumar, M.R.; Arunagirinathan, N.; Al-Dhabi, N.A.; Valan Arasu, M.; Ignacimuthu, S.R.; Indinavir, T. Dolutegravir, and Etravirine against main protease and RNA-dependent RNA polymerase of SARS-CoV-2: A molecular docking and drug repurposing approach. J. Infect. Public Health 2020, 13, 1856–1861. [Google Scholar] [CrossRef]
- Chien, M.; Anderson, T.K.; Jockusch, S.; Tao, C.; Li, X.; Kumar, S.; Russo, J.J.; Kirchdoerfer, R.N.; Ju, J. Nucleotide Analogues as Inhibitors of SARS-CoV-2 Polymerase, a Key Drug Target for COVID-19. J. Proteome Res. 2020, 19, 4690–4697. [Google Scholar] [CrossRef] [PubMed]
- Ayerdi, O.; Puerta, T.; Clavo, P.; Vera, M.; Ballesteros, J.; Fuentes, M.E. Preventive Efficacy of Tenofovir/Emtricitabine Against Severe Acute Respiratory Syndrome Coronavirus 2 Among Pre-Exposure Prophylaxis Users. Open Forum Infect Dis. 2020, 7, ofaa455. [Google Scholar] [CrossRef]
- Peele, K.A.; Durthi, C.P.; Srihansa, T.; Krupanidhi, S.; Ayyagari, V.S.; Babu, D.J. Molecular docking and dynamic simulations for antiviral compounds against SARS-CoV-2: A computational study. Inform. Med. Unlocked 2020, 19, 100345. [Google Scholar] [CrossRef] [PubMed]
- Mostafa, M.A. Role of Zidovudine and Candesartan in the Novel SARS-CoV-2 Treatment Trials; Theoretical Study. AIJR Prepr. 2020, 30. [Google Scholar] [CrossRef]
- Fintelman-Rodrigues, N.; Sacramento, C.Q.; Lima, C.R.; da Silva, F.S.; Ferreira, A.C.; Mattos, M.; de Freitas, C.S.; Soares, V.C.; Dias, S.D.S.G.; Temerozo, J.R.; et al. Atazanavir, Alone or in Combination with Ritonavir, Inhibits SARS-CoV-2 Replication and Proinflammatory Cytokine Production. Antimicrob. Agents Chemother. 2020, 64. [Google Scholar] [CrossRef]
- Panagopoulos, P.; Petrakis, V.; Panopoulou, M.; Trypsianis, G.; Penlioglou, T.; Pnevmatikos, I.; Papazoglou, D. Lopinavir/ritonavir as a third agent in the antiviral regimen for SARS-CoV-2 infection. J. Chemother. 2020, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, M.; Schroeder, S.; Kleine-Weber, H.; Müller, M.A.; Drosten, C.; Pöhlmann, S. Nafamostat Mesylate Blocks Activation of SARS-CoV-2: New Treatment Option for COVID-19. Antimicrob. Agents Chemother. 2020, 64. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shytaj, I.L.; Fares, M.; Lucic, B.; Gallucci, L.; Tolba, M.M.; Zimmermann, L.; Ayoub, A.T.; Cortese, M.; Neufeldt, C.J.; Laketa, V.; et al. The FDA-approved drug cobicistat synergizes with remdesivir to inhibit SARS-CoV-2 replication. bioRxiv 2021. [Google Scholar] [CrossRef]
- Choy, K.T.; Wong, A.Y.L.; Kaewpreedee, P.; Sia, S.F.; Chen, D.; Hui, K.P.Y. Remdesivir, lopinavir, emetine, and homoharringtonine inhibit SARSCoV-2 replication in vitro. Antiviral. Res. 2020, 178, 104786. [Google Scholar] [CrossRef]
- Qiao, Z.; Zhang, H.; Ji, H.-F.; Chen, Q. Computational View toward the Inhibition of SARS-CoV-2 Spike Glycoprotein and the 3CL Protease. Computation 2020, 8, 53. [Google Scholar] [CrossRef]
- Bharadwaj, S.; Azhar, E.I.; Kamal, M.A.; Bajrai, L.H.; Dubey, A.; Jha, K.; Yadava, U.; Kang, S.G.; Dwivedi, V.D. SARS-CoV-2 Mpro inhibitors: Identification of anti-SARS-CoV-2 Mpro compounds from FDA approved drugs. J. Biomol. Struct. Dyn. 2020. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Cao, R.; Xu, M.; Wang, X.; Zhang, H.; Hu, H.; Li, Y.; Hu, Z.; Zhong, W.; Wang, M. Hydroxychloroquine, a less toxic derivative of chloroquine, is effective in inhibiting SARS-CoV-2 infection in vitro. Cell Discov. 2020, 6, 16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raposo, C.; Nunes, A.K.D.; Luna, R.L.D.; Araújo, S.M.D.; da Cruz-Höfling, M.A.; Peixoto, C.A. Sildenafil (Viagra) Protective Effects on Neuroinflammation: The Role of iNOS/NO System in an Inflammatory Demyelination Model. Mediat. Inflamm. 2013, 2013, 321460. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nunes, A.K.S.; Rapôso, C.; Rocha, S.W.S.; de Sousa Barbosa, K.P.; de Almeida Luna, R.L.; da Cruz-Höfling, M.A.; Peixoto, C.A. Involvement of AMPK, IKβα-NFκB and eNOS in the sildenafil anti-inflammatory mechanism in a demyelination model. Brain Res. 2015, 1627, 119–133. [Google Scholar] [CrossRef] [PubMed]
- Morais, I.J.; Polveiro, R.C.; Souza, G.M.; Bortolin, D.I.; Sassaki, F.T.; Lima, A.T.M. The global population of SARS-CoV-2 is composed of six major subtypes. Sci. Rep. 2020, 10, 18289. [Google Scholar] [CrossRef] [PubMed]
- Clososki, G.C.; Soldi, R.A.; da Silva, R.M.; Guaratini, T.; Lopes, J.N.C.; Pereira, P.R.R.; Lopes, J.L.C.; dos Santos, T.; Martins, R.B.; Costa, C.S.; et al. Tenofovir Disoproxil Fumarate: New Chemical Developments and Encouraging in vitro Biological Results for SARS-CoV-2. J. Braz. Chem. Soc. 2020, 31, 1552–1556. [Google Scholar] [CrossRef]
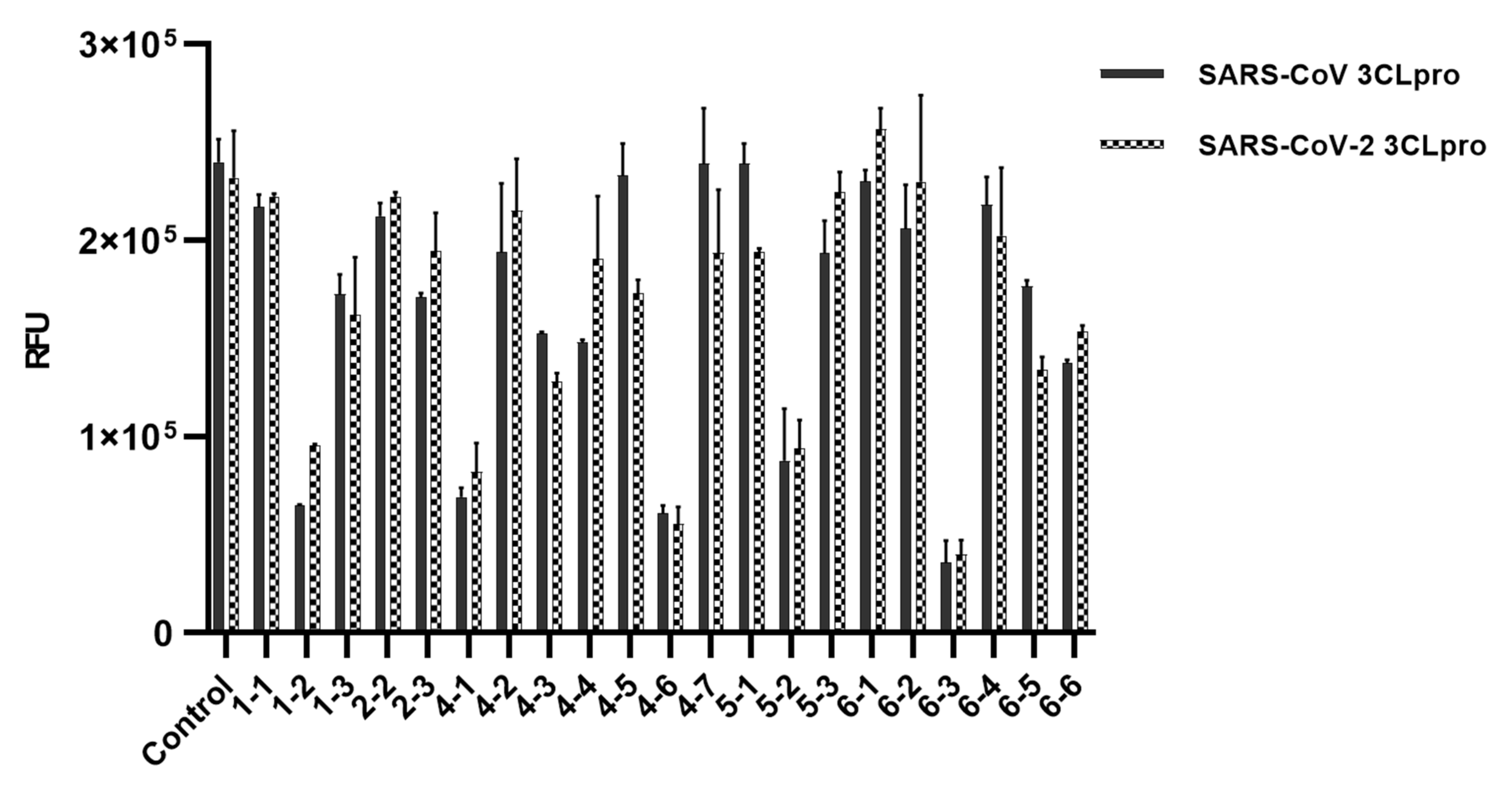
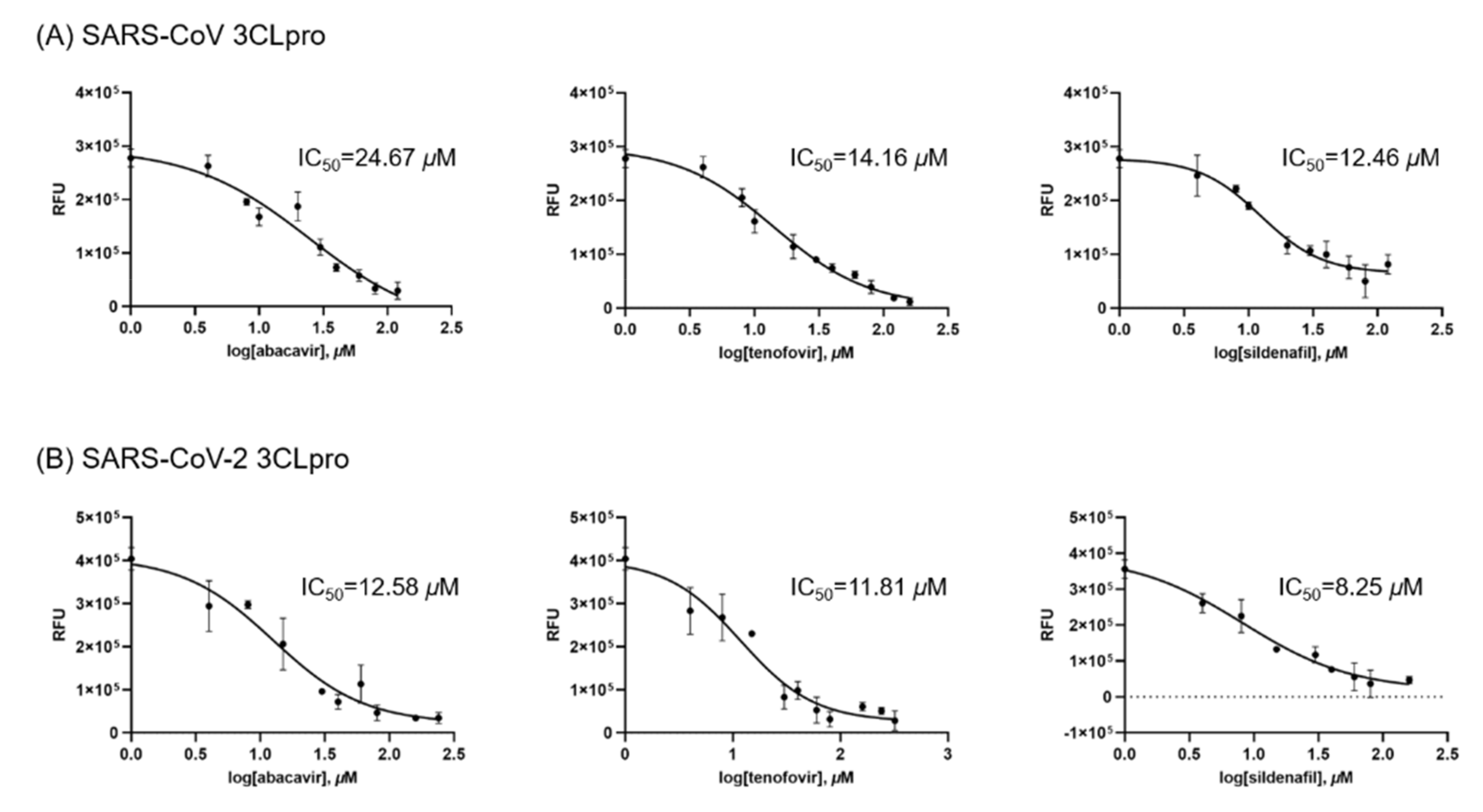
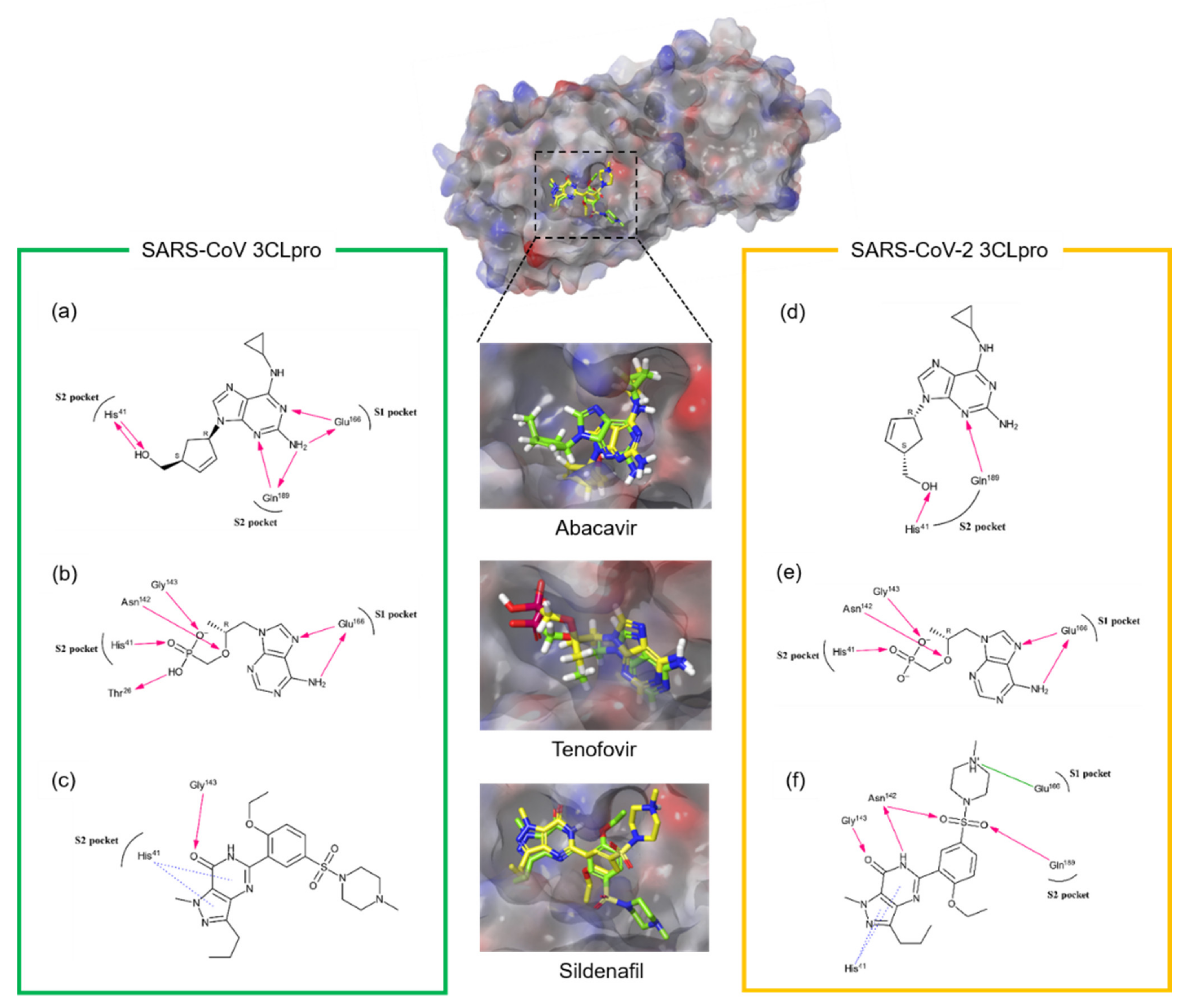
| No | Name of Compound | Molecular Weight | Molecular Formula | CAS Number | Reference |
|---|---|---|---|---|---|
| 1-1 | Acyclovir | 225.20 | C8H11N5O3 | 59277-89-3 | [23] |
| 1-2 | Cidofovir | 279.19 | C8H14N3O6P xH2O | 113852-37-2 | [24] |
| 1-3 | Ganciclovir | 255.23 | C9H13N5O4 | 82410-32-0 | [25] |
| 2-1 | Favipiravir | 157.10 | C5H4FN3O2 | 259793-96-9 | [26] |
| 2-2 | Remdesivir | 291.27 | C12H13N5O4 | 1809249-37-3 | [27,28] |
| 2-3 | Sofosbuvir | 529.45 | C22H29FN3O9P | 82-93-9 | [29] |
| 3-1 | Raltegravir | 482.51 | C20H20FKN6O5 | 871038-72-1 | [30] |
| 4-1 | Abacavir | 225.20 | C8H11N5O3 | 188062-50-2 | [31] |
| 4-2 | Emtricitabine | 247.25 | C8H10FN3O3S | 143491-57-0 | [32] |
| 4-3 | Entecavir | 277.28 | C12H15N5O3 | 142217-69-4 | [33] |
| 4-4 | Lamivudine | 229.26 | C8H11N3O3S | 134678-17-4 | [31] |
| 4-5 | Ribavirin | 244.20 | C8H12N4O5 | 36791-04-5 | [31] |
| 4-6 | Tenofovir | 305.23 | C9H14N5O4P H2O | 206184-49-8 | [32] |
| 4-7 | Zidovudine | 267.24 | C10H13N5O4 | 30516-87-1 | [34] |
| 5-1 | Darunavir | 547.66 | C27H37N3O7S | 206361-99-1 | [35] |
| 5-2 | Indinavir | 613.79 | C36H49N5O8S | 180683-37-8 | [30] |
| 5-3 | Lopinavir | 628.80 | C37H48N4O5 | 192725-17-0 | [36] |
| 5-4 | Nafamostat mesylate | 539.58 | C19H17N5O2 2CH4O3S | 82956-11-4 | [37] |
| 6-1 | Cobicistat | 776.0 | C40H53N7O5S2 | 1004316-88-4 | [38] |
| 6-2 | Ritonavir | 720.94 | C37H48N6O5S2 | 155213-67-5 | [27,39] |
| 6-3 | Sildenafil | 666.70 | C22H30N6O4S C6H8O7 | 171599-83-0 | [40] |
| 6-4 | Tadalafil | 389.40 | C22H19N3O4 | 171596-29-5 | [41] |
| 6-5 | Chloroquine | 515.86 | C18H26ClN3 2H3PO4 | 50-63-5 | [42] |
| 6-6 | Hydroxychloroquine | 433.95 | C18H26ClN3O H2SO4 | 747-36-4 | [42] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jo, S.; Kim, S.; Yoo, J.; Kim, M.-S.; Shin, D.H. A Study of 3CLpros as Promising Targets against SARS-CoV and SARS-CoV-2. Microorganisms 2021, 9, 756. https://doi.org/10.3390/microorganisms9040756
Jo S, Kim S, Yoo J, Kim M-S, Shin DH. A Study of 3CLpros as Promising Targets against SARS-CoV and SARS-CoV-2. Microorganisms. 2021; 9(4):756. https://doi.org/10.3390/microorganisms9040756
Chicago/Turabian StyleJo, Seri, Suwon Kim, Jahyun Yoo, Mi-Sun Kim, and Dong Hae Shin. 2021. "A Study of 3CLpros as Promising Targets against SARS-CoV and SARS-CoV-2" Microorganisms 9, no. 4: 756. https://doi.org/10.3390/microorganisms9040756
APA StyleJo, S., Kim, S., Yoo, J., Kim, M.-S., & Shin, D. H. (2021). A Study of 3CLpros as Promising Targets against SARS-CoV and SARS-CoV-2. Microorganisms, 9(4), 756. https://doi.org/10.3390/microorganisms9040756





