Early Post-Prandial Regulation of Protein Expression in the Midgut of Chagas Disease Vector Rhodnius prolixus Highlights New Potential Targets for Vector Control Strategy
Abstract
:1. Introduction
2. Materials and Methods
2.1. Samples Preparation
2.2. Dissection of the Insect and Tissues Preparation
2.3. Proteins Extraction
2.4. Sample Preparation Prior to Liquid Chromatography Tandem Mass Spectrometry
2.5. Mass Spectrometric Data Analysis
2.6. Functional Characterization and Protein Classification
2.7. Western Blotting
3. Results
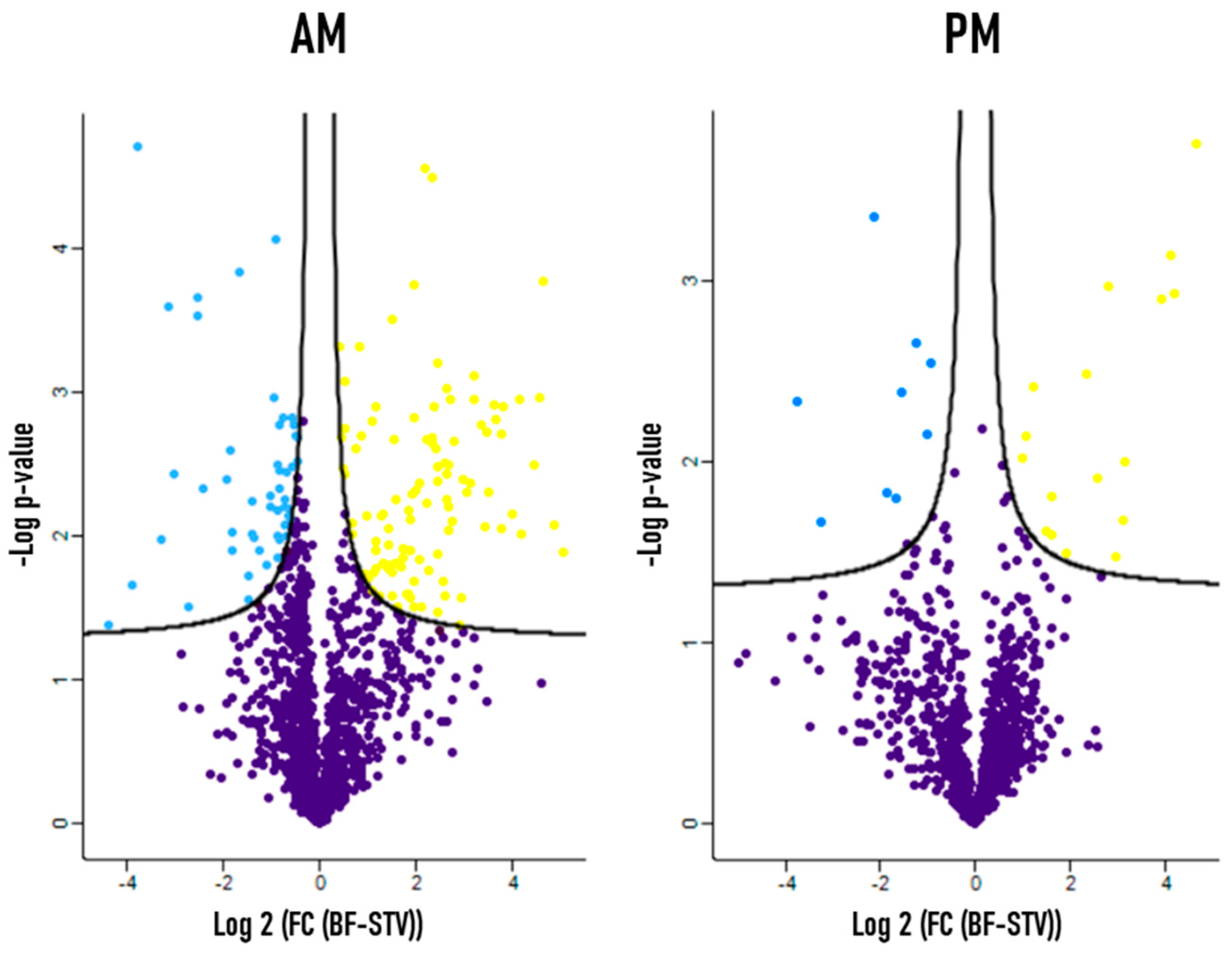
3.1. Differential Protein Expression between Starved and Blood Fed Conditions
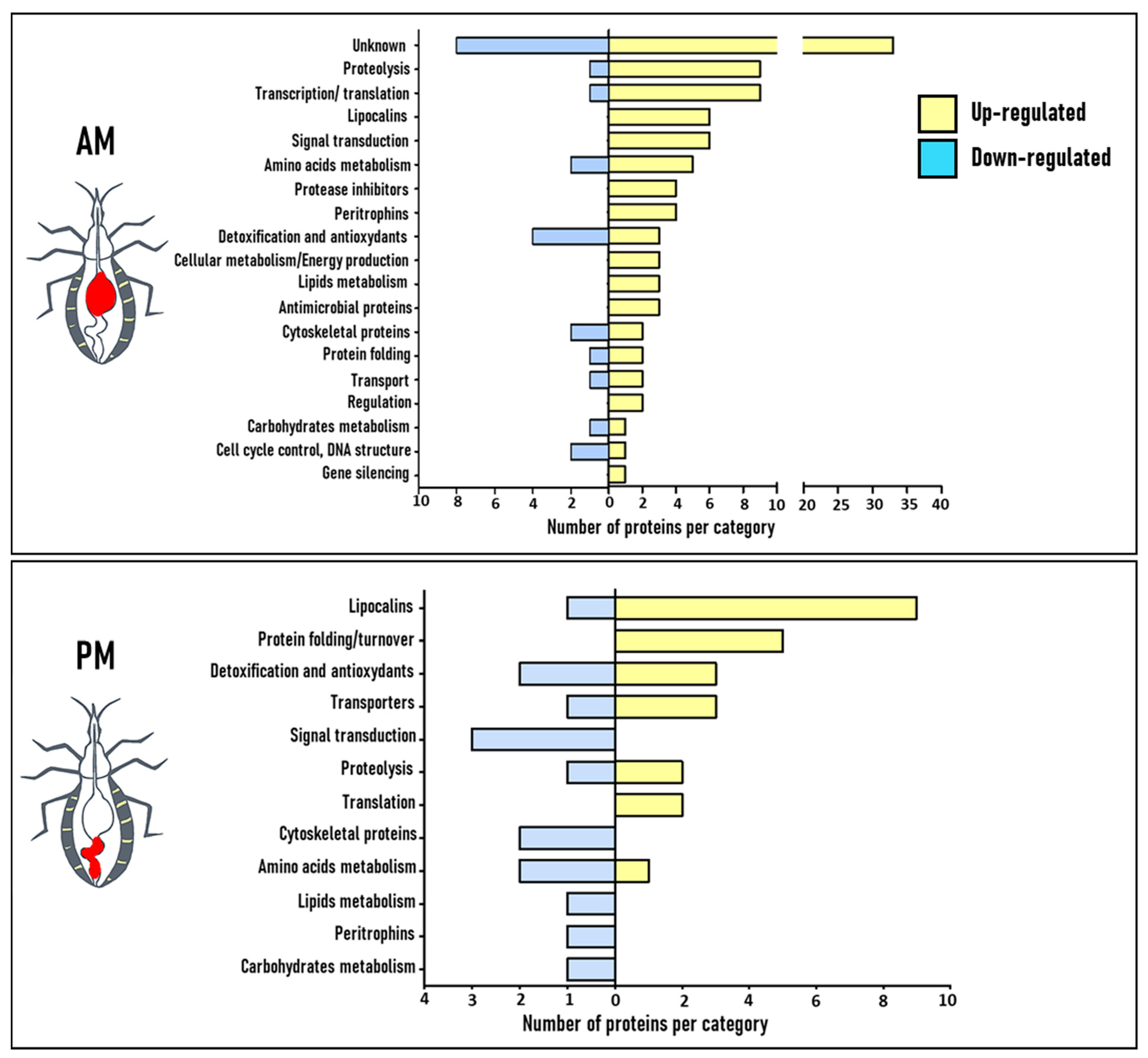
3.2. Functional Annotation of Blood Regulated Proteins
3.3. Validation by Western Blot of Differentially Expressed Proteins Candidates
4. Discussion
4.1. A Large Blood Meal Has Limited Effect on the Dynamic of Protein Expression in R. prolixus Midgut during the First Hours after Feeding
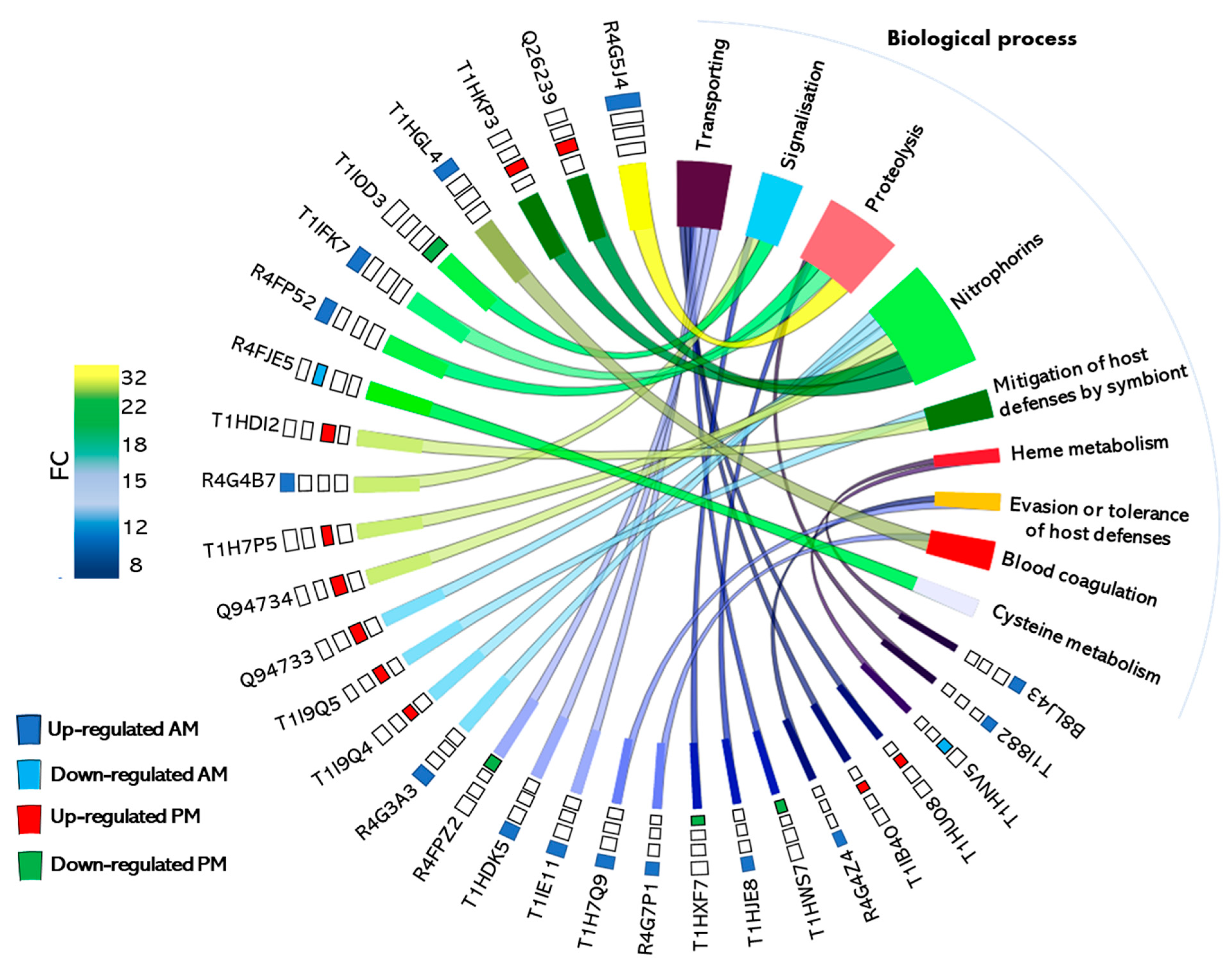
4.2. Detailed Analysis of the Differentially Expressed Proteins and Their Physiological Involvement in Blood Digestion
4.2.1. The Vast Majority of the Differentially Expressed Proteins in the Midgut in Response to Blood Intake Are Involved in a Physiological Role Related to Blood Processing
- Proteases
- Protease Inhibitors
- Amino Acids Metabolism
- Lipocalins
- Proteins involved in transcription and translation machinery
4.2.2. A Timid Stress Response Is Induced in the Midgut of R. prolixus in the Early Hours Post Blood Feeding
- Oxidative and heat stress response proteins
- Innate Immunity related proteins
4.3. New Insights about Unknown Proteins
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Barrozo, R.B. Food recognition in hematophagous insects. Curr. Opin. Insect Sci. 2019, 34, 55–60. [Google Scholar] [CrossRef]
- Hurd, H. Parasite-Modified Vector Behavior. Encycl. Anim. Behav. 2009, 628–631. [Google Scholar] [CrossRef]
- Garcia, E.S.; Genta, F.A.; De Azambuja, P.; Schaub, G.A. Interactions between intestinal compounds of triatomines and Trypanosoma cruzi. Trends Parasitol. 2010, 26, 499–505. [Google Scholar] [CrossRef] [PubMed]
- World Health Organisation (WHO). Chagas Disease (Also Known as American Trypanosomiasis). 2020. Available online: https://www.who.int/news-room/fact-sheets/detail/chagas-disease-(american-trypanosomiasis) (accessed on 4 March 2021).
- Rassi, A.J.; Rassi, A.; Marin-neto, J.A. Chagas disease. Lancet 2010, 375, 1388–1402. [Google Scholar] [CrossRef]
- Kollien, A.H.; Schaub, G.A. The Development of Trypanosoma cruzi in Triatominae. Parasitol. Today 2000, 16, 381–387. [Google Scholar] [CrossRef]
- Zeledn, R.; Rica, U.D.C.; Jose, S.; Rica, C. Chagas Disease: An Ecological Appraisal with Special Emphasis on Its Insect Vectors. Ann. Rev. Entomol 1981, 26, 101–133. [Google Scholar] [CrossRef] [PubMed]
- Fronza, G.; Roca-Acevedo, G.; Mougabure-Cueto, G.A.; Sierra, I.; Capriotti, N.; Toloza, A.C. Insecticide Resistance Mechanisms in Triatoma infestans (Reduviidae: Triatominae): The Putative Role of Enhanced Detoxification and Knockdown Resistance (kdr) Allele in a Resistant Hotspot From the Argentine Chaco. J. Med. Entomol. 2020, 57, 837–844. [Google Scholar] [CrossRef] [PubMed]
- Germano, M.D.; Santo-Orihuela, P.; Roca-Acevedo, G.; Toloza, A.C.; Vassena, C.; Picollo, M.I.; Mougabure-Cueto, G. Scientific evidence of three different insecticide-resistant profiles in Triatoma infestans (Hemiptera: Reduviidae) populations from Argentina and Bolivia. J. Med. Entomol. 2012, 49, 1355–1360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mougabure-Cueto, G.; Picollo, M.I. Insecticide resistance in vector Chagas disease: Evolution, mechanisms and management. Acta Trop. 2015, 149, 70–85. [Google Scholar] [CrossRef]
- Balczun, C.; Pausch, J.K.; Schaub, G.A. Blood digestion in triatomines—A review. Mitt. Dtsch. Ges. Allg. Angew. Ent. 2012, 18, 331–334. [Google Scholar]
- Nunes-da-Fonseca, R.; Berni, M.; Tobias-dos-Santos, V.; Pane, A.; Araujo, H.M. Rhodnius prolixus: From classical physiology to modern developmental biology. Genesis 2017, 55, e22995. [Google Scholar] [CrossRef] [Green Version]
- Guarneri, A.A.; Lorenzo, M.G. Triatomine physiology in the context of trypanosome infection. J. Insect Physiol. 2017, 97, 66–76. [Google Scholar] [CrossRef]
- de Azambuja, P.; Guimarães, J.A.; Garcia, E.S. Haemolytic factor from the crop of Rhodnius prolixus: Evidence and partial characterization. J. Insect Physiol. 1983, 29, 833–837. [Google Scholar] [CrossRef]
- Friedrich, T.; Kroger, B.; Bialojan, S.; Lemaire, H.G.; Hoffken, H.W.; Reuschenbach, P.; Otte, M.; Dodt, J. A Kazal-type inhibitor with thrombin specificity from Rhodnius prolixus. J. Biol. Chem. 1993, 268, 16216–16222. [Google Scholar] [CrossRef]
- Araujo, R.N.; Campos, I.T.N.; Tanaka, A.S.; Santos, A.; Gontijo, N.F.; Lehane, M.J.; Pereira, M.H. Brasiliensin: A novel intestinal thrombin inhibitor from Triatoma brasiliensis (Hemiptera: Reduviidae) with an important role in blood intake. Int. J. Parasitol. 2007, 37, 1351–1358. [Google Scholar] [CrossRef] [Green Version]
- Campos, I.T.N.; Amino, R.; Sampaio, C.A.M.; Auerswald, E.A.; Friedrich, T.; Lemaire, H.G.; Schenkman, S.; Tanaka, A.S. Infestin, a thrombin inhibitor presents in Triatoma infestans midgut, a Chagas’ disease vector: Gene cloning, expression and characterization of the inhibitor. Insect Biochem. Mol. Biol. 2002, 32, 991–997. [Google Scholar] [CrossRef]
- van de Locht, A.; Lamba, D.; Bauer, M.; Huber, R.; Friedrich, T.; Kröger, B.; Höffken, W.; Bode, W. Two heads are better than one: Crystal structure of the insect derived double domain Kazal inhibitor rhodniin in complex with thrombin. EMBO J. 1995, 14, 5149–5157. [Google Scholar] [CrossRef] [PubMed]
- Henriques, B.S.; Gomes, B.; Oliveira, P.L.; Garcia, E.D.S.; Azambuja, P.; Genta, F.A. Characterization of the Temporal Pattern of Blood Protein Digestion in Rhodnius prolixus: First Description of Early and Late Gut Cathepsins. Front. Physiol. 2021, 11, 509310. [Google Scholar] [CrossRef] [PubMed]
- Ouali, R.; de Brito, K.C.V.; Salmon, D.; Bousbata, S. High-Throughput Identification of the Rhodnius prolixus Midgut Proteome Unravels a Sophisticated Hematophagic Machinery. Proteomes 2020, 8, 16. [Google Scholar] [CrossRef] [PubMed]
- Eichler, S.; Schaub, G.A. Development of symbionts in triatomine bugs and the effects of infections with trypanosomatids. Exp. Parasitol. 2002, 100, 17–27. [Google Scholar] [CrossRef] [PubMed]
- Kollien, A.H.; Schaub, G.A. Trypanosoma cruzi in the rectum of the bug Triatoma infestans: Effects of blood ingestion by the starved vector. Am. J. Trop. Med. Hyg. 1998, 59, 166–170. [Google Scholar] [CrossRef] [Green Version]
- de Almeida Dias, F.; Guerra, B.; Vieira, L.R.; Perdomo, H.D.; Gandara, A.C.P.; do Amaral, R.J.V.; Vollú, R.E.; Gomes, S.A.O.; Lara, F.A.; Sorgine, M.H.F.; et al. Monitoring of the Parasite Load in the Digestive Tract of Rhodnius prolixus by Combined qPCR Analysis and Imaging Techniques Provides New Insights into the Trypanosome Life Cycle. PLoS Negl. Trop. Dis. 2015, 9, e0004186. [Google Scholar]
- Ferreira, R.C.; Kessler, R.L.; Lorenzo, M.G.; Paim, R.M.M.; Ferreira, L.D.L.; Probst, C.M.; Alves-Silva, J.; Guarneri, A.A. Colonization of Rhodnius prolixus gut by Trypanosoma cruzi involves an extensive parasite killing. Parasitology 2016, 143, 434–443. [Google Scholar] [CrossRef] [PubMed]
- Gonçalves, C.S.; Ávila, A.R.; De Souza, W.; Motta, M.C.M.; Cavalcanti, D.P. Revisiting the Trypanosoma cruzi metacyclogenesis: Morphological and ultrastructural analyses during cell differentiation. Parasit. Vectors 2018, 11, 83. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Neuhauser, N.; Michalski, A.; Scheltema, R.A.; Olsen, J.V.; Mann, M. Andromeda: A Peptide Search Engine Integrated into the MaxQuant Environment. J. Proteome Res. 2011, 10, 1794–1805. [Google Scholar]
- Mi, H.; Muruganujan, A.; Casagrande, J.T.; Thomas, P.D. Large-scale gene function analysis with PANTHER classification system. Nat. Protoc. 2013, 8, 1551–1566. [Google Scholar] [CrossRef] [PubMed]
- Cázares-Raga, F.E.; Chávez-Munguía, B.; González-Calixto, C.; Ochoa-Franco, A.P.; Gawinowicz, M.A.; Rodríguez, M.H.; Hernández-Hernández, F.C. Morphological and proteomic characterization of midgut of the malaria vector Anopheles albimanus at early time after a blood feeding. J. Proteom. 2014, 111, 100–112. [Google Scholar] [CrossRef] [PubMed]
- Dana, A.N.; Hong, Y.S.; Kern, M.K.; Hillenmeyer, M.E.; Harker, B.W.; Lobo, N.F.; Hogan, J.R.; Romans, P.; Collins, F.H. Gene expression patterns associated with blood-feeding in the malaria mosquito Anopheles gambiae. BMC Genom. 2005, 6, 5. [Google Scholar] [CrossRef] [Green Version]
- Prévot, G.I.; Laurent-Winter, C.; Rodhain, F.; Bourgouin, C. Sex-specific and blood meal-induced proteins of Anopheles gambiae midguts: Analysis by two-dimensional gel electrophoresis. Malar. J. 2003, 2, 1–17. [Google Scholar] [CrossRef]
- Dienge, H.; Boots, M.; Higashihara, J.; Okada, T.; Kato, K.; Satho, T.; Miake, F.; Eshita, Y. Effects of blood and virus-infected blood on protein expression in the midgut of the dengue vector Aedes albopictus. Med. Vet. Entomol. 2007, 21, 278–283. [Google Scholar] [CrossRef]
- Bonizzoni, M.; Dunn, W.A.; Campbell, C.L.; Olson, K.E.; Dimon, M.T.; Marinotti, O.; James, A.A. RNA-seq analyses of blood-induced changes in gene expression in the mosquito vector species, Aedes aegypti. BMC Genom. 2011, 12, 82. [Google Scholar] [CrossRef] [Green Version]
- Dostálová, A.; Votýpka, J.; Favreau, A.J.; Barbian, K.D.; Volf, P.; Valenzuela, J.G.; Jochim, R.C. The midgut transcriptome of Phlebotomus (Larroussius) perniciosus, a vector of Leishmania infantum: Comparison of sugar fed and blood fed sand flies. BMC Genom. 2011, 12, 223. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schwarz, A.; Tenzer, S.; Hackenberg, M.; Erhart, J.; Gerhold-Ay, A.; Mazur, J.; Kuharev, J.; Ribeiro, J.M.C.; Kotsyfakis, M. A Systems Level Analysis Reveals Transcriptomic and Proteomic Complexity in Ixodes ricinus Midgut and Salivary Glands During Early Attachment and Feeding. Mol. Cell. Proteomics 2014, 13, 2725–2735. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rudenko, N.; Golovchenko, M.; Edwards, M.J. Differential Expression of Ixodes ricinus Tick Genes Induced by Blood Feeding or Borrelia burgdorferi Infection. J. Med. Entomol. 2005, 1, 36–41. [Google Scholar] [CrossRef] [Green Version]
- Oleaga, A.; Obolo-Mvoulouga, P.; Manzano-Román, R.; Pérez-Sánchez, R. Midgut proteome of an argasid tick, Ornithodoros erraticus: A comparison between unfed and engorged females. Parasit. Vectors 2015, 8, 525. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oleaga, A.; Obolo-mvoulouga, P.; Manzano-román, R.; Pérez-sánchez, R. A proteomic insight into the midgut proteome of Ornithodoros moubata females reveals novel information on blood digestion in argasid ticks. Parasit. Vectors 2017, 10, 40–52. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Terra, W.R.; Dias, R.O.; Ferreira, C. Recruited lysosomal enzymes as major digestive enzymes in insects. Biochem. Soc. Trans. 2019, 47, 615–623. [Google Scholar] [CrossRef] [PubMed]
- Henriques, B.S.; Gomes, B.; da Costa, S.G.; da Silva Moraes, C.; Mesquita, R.D.; Dillon, V.M.; de Souza Garcia, E.; Azambuja, P.; Dillon, R.J.; Genta, F.A. Genome wide mapping of peptidases in Rhodnius prolixus: Identification of protease gene duplications, horizontally transferred proteases and analysis of peptidase a1 structures, with considerations on their role in the evolution of hematophagy in triatomi. Front. Physiol. 2017, 8, 1051. [Google Scholar] [CrossRef] [Green Version]
- Ribeiro, J.M.C.; Genta, F.A.; Sorgine, M.H.F.; Logullo, R.; Mesquita, R.D.; Paiva-Silva, G.O.; Majerowicz, D.; Medeiros, M.; Koerich, L.; Terra, W.R.; et al. An Insight into the Transcriptome of the Digestive Tract of the Bloodsucking Bug, Rhodnius prolixus. PLoS Negl. Trop. Dis. 2014, 8, e2594. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Terra, W.R.; Ferreira, C.; Garcia, E.S. Origin, distribution, properties and functions of the major Rhodnius prolixus midgut hydrolases. Insect Biochem. 1988, 18, 423–434. [Google Scholar] [CrossRef]
- Borges, E.C.; Machado, E.M.M.; Garcia, E.S.; Azambuja, P. Trypanosoma cruzi: Effects of infection on cathepsin D activity in the midgut of Rhodnius prolixus. Exp. Parasitol. 2006, 112, 130–133. [Google Scholar] [CrossRef] [PubMed]
- Buarque, D.S.; Braz, G.R.C.; Martins, R.M.; Tanaka-Azevedo, A.M.; Gomes, C.M.; Oliveira, F.A.A.; Schenkman, S.; Tanaka, A.S. Differential Expression Profiles in the Midgut of Triatoma infestans Infected with Trypanosoma cruzi. PLoS ONE 2013, 8, e61203. [Google Scholar] [CrossRef] [Green Version]
- Balczun, C.; Siemanowski, J.; Pausch, J.K.; Helling, S.; Marcus, K.; Stephan, C.; Meyer, H.E.; Schneider, T.; Cizmowski, C.; Oldenburg, M.; et al. Intestinal aspartate proteases TiCatD and TiCatD2 of the haematophagous bug Triatoma infestans (Reduviidae): Sequence characterisation, expression pattern and characterisation of proteolytic activity. Insect Biochem. Mol. Biol. 2012, 42, 240–250. [Google Scholar] [CrossRef] [PubMed]
- Waniek, P.J.; Pacheco Costa, J.E.; Jansen, A.M.; Costa, J.; Araújo, C.A.C. Cathepsin L of Triatoma brasiliensis (Reduviidae, Triatominae): Sequence characterization, expression pattern and zymography. J. Insect Physiol. 2012, 58, 178–187. [Google Scholar] [CrossRef] [Green Version]
- Kollien, A.H.; Waniek, P.J.; Nisbet, A.J.; Billingsley, P.F.; Schaub, G.A. Activity and sequence characterization of two cysteine proteases in the digestive tract of the reduviid bug Triatoma infestans. Insect Mol. Biol. 2004, 13, 569–579. [Google Scholar] [CrossRef]
- Waniek, P.J.; Araújo, C.A.C.; Momoli, M.M.; Azambuja, P.; Jansen, A.M.; Genta, F.A. Serine carboxypeptidases of Triatoma brasiliensis (Hemiptera, Reduviidae): Sequence characterization, expression pattern and activity localization. J. Insect Physiol. 2014, 63, 9–20. [Google Scholar] [CrossRef]
- Francischetti, I.M.B.; Mather, T.N.; Ribeiro, J.M.C. Cloning of a salivary gland metalloprotease and characterization of gelatinase and fibrin(ogen)lytic activities in the saliva of the Lyme disease tick vector Ixodes scapularis. Biochem. Biophys. Res. Commun. 2003, 305, 869–875. [Google Scholar] [CrossRef] [Green Version]
- Guo, X.; Booth, C.J.; Paley, M.A.; Wang, X.; DePonte, K.; Fikrig, E.; Narasimhan, S.; Montgomery, R.R. Inhibition of neutrophil function by two tick salivary proteins. Infect. Immun. 2009, 77, 2320–2329. [Google Scholar] [CrossRef] [Green Version]
- Barnard, A.C.; Nijhof, A.M.; Gaspar, A.R.M.; Neitz, A.W.H.; Jongejan, F.; Maritz-Olivier, C. Expression profiling, gene silencing and transcriptional networking of metzincin metalloproteases in the cattle tick, Rhipicephalus (Boophilus) microplus. Vet. Parasitol. 2012, 186, 403–414. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feder, D.; Salles, J.M.; Garcia, E.S.; Azambuja, P. Haemolymph and Fat Body Metallo-protease Associated with Enterobacter cloacae Infection in the Bloodsucking Insect, Rhodnius prolixus. Mem. Inst. Oswaldo Cruz 1998, 93, 823–826. [Google Scholar] [CrossRef] [Green Version]
- Feder, D.; Gomes, S.A.O.; Garcia, E.S.; Azambuja, P. Metalloproteases in Trypanosoma rangeli-infected Rhodnius prolixus. Mem. Inst. Oswaldo Cruz 1999, 94, 771–777. [Google Scholar] [CrossRef] [Green Version]
- Santiago, P.B.; De Araújo, C.N.; Motta, F.N.; Praça, Y.R.; Charneau, S.; Bastos, I.M.D.; Santana, J.M. Proteases of haematophagous arthropod vectors are involved in blood-feeding, yolk formation and immunity—A review. Parasit. Vectors 2017, 10, 79. [Google Scholar] [CrossRef] [Green Version]
- Gubb, D.; Sanz-Parra, A.; Barcena, L.; Troxler, L.; Fullaondo, A. Protease inhibitors and proteolytic signalling cascades in insects. Enferm. Infecc. Microbiol. Clin. 2010, 28, 1749–1759. [Google Scholar] [CrossRef]
- Ledizet, M.; Harrison, L.M.; Koski, R.A.; Cappello, M. Discovery and pre-clinical development of antithrombotics from hematophagous invertebrates. Curr. Med. Chem. Cardiovasc. Hematol. Agents 2005, 3, 1–10. [Google Scholar] [CrossRef]
- Meiser, C.K.; Piechura, H.; Werner, T.; Dittmeyer-Schäfer, S.; Meyer, H.E.; Warscheid, B.; Schaub, G.A.; Balczun, C. Kazal-type inhibitors in the stomach of Panstrongylus megistus (Triatominae, Reduviidae). Insect Biochem. Mol. Biol. 2010, 40, 345–353. [Google Scholar] [CrossRef]
- Noeske-Jungblut, C.; Haendler, B.; Donner, P.; Alagon, A.; Possani, L.; Schleuning, W.D. Triabin, a highly potent exosite inhibitor of thrombin. J. Biol. Chem. 1995, 270, 28629–28634. [Google Scholar] [CrossRef] [Green Version]
- Soares, T.S.; Buarque, D.S.; Queiroz, B.R.; Gomes, C.M.; Braz, G.R.C.; Araújo, R.N.; Pereira, M.H.; Guarneri, A.A.; Tanaka, A.S. A Kazal-type inhibitor is modulated by Trypanosoma cruzi to control microbiota inside the anterior midgut of Rhodnius prolixus. Biochimie 2015, 112, 41–48. [Google Scholar] [CrossRef] [PubMed]
- Fuentes-Prior, P.; Noeske-Jungblut, C.; Donner, P.; Schleuning, W.D.; Huber, R.; Bode, W. Structure of the thrombin complex with triabin, a lipocalin-like exosite-binding inhibitor derived from a triatomine bug. Proc. Natl. Acad. Sci. USA 1997, 94, 11845–11850. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sterkel, M.; Oliveira, J.H.M.; Bottino, V.; Paiva-Silva, G.O.; Oliveira, P.L. The Dose Makes the Poison: Nutritional Overload Determines the Life Traits of Blood-Feeding Arthropods. Trends Parasitol. 2017, 33, 633–644. [Google Scholar] [CrossRef] [PubMed]
- Sterkel, M.; Perdomo, H.D.; Guizzo, M.G.; Dias, F.A.; Sorgine, M.H.F.; Oliveira, P.L.; Sterkel, M.; Perdomo, H.D.; Guizzo, M.G.; Barletta, A.B.F.; et al. Tyrosine Detoxification Is an Essential Trait in the Life History of Blood-Feeding Arthropods. Curr. Biol. 2016, 26, 2188–2193. [Google Scholar] [CrossRef] [Green Version]
- Sterkel, M.; Oliveira, P.L. Developmental roles of tyrosine metabolism enzymes in the blood- sucking insect Rhodnius prolixus. Proc. R. Soc. B 2017, 284, 20162607. [Google Scholar] [CrossRef] [Green Version]
- Sterkel, M.; Haines, L.R.; Casas-Sánchez, A.; Owino Adung’a, V.; Vionette-Amaral, R.J.; Quek, S.; Rose, C.; Silva dos Santos, M.; García Escude, N.; Ismail, H.M.; et al. Repurposing the orphan drug nitisinone to control the transmission of African trypanosomiasis. PLoS Biol. 2021, 19, e3000796. [Google Scholar] [CrossRef]
- Flower, D.R. The lipocalin protein family: Structure and function. Biochem. J. 1996, 318, 1–14. [Google Scholar] [CrossRef]
- Francischetti, I.M.B.; Ribeiro, J.M.C.; Champagne, D.; Andersen, J. Purification, Cloning, Expression, and Mechanism of Action of a Novel Platelet Aggregation Inhibitor from the Salivary Gland of the Blood-sucking Bug, Rhodnius prolixus. J. Biol. Chem. 2000, 275, 12639–12650. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Champagne, D.E. Antihemostatic Molecules from Saliva of Blood-Feeding Arthropods. Pathophysiol. Haemost. Thromb. 2005, 30602, 221–227. [Google Scholar] [CrossRef]
- Araujo, R.N.; Soares, A.C.; Paim, R.M.M.; Gontijo, N.F.; Gontijo, A.F.; Lehane, M.J.; Pereira, M.H. The role of salivary nitrophorins in the ingestion of blood by the triatomine bug Rhodnius prolixus (Reduviidae: Triatominae). Insect Biochem. Mol. Biol. 2009, 39, 83–89. [Google Scholar] [CrossRef] [PubMed]
- Andersen, J.; Gudderra, N.; Francischetti, I.M.B.; Ribeiro, J.M.C. The role of salivary lipocalins in blood feeding by Rhodnius prolixus. Arch. Insect Biochem. Physiol. 2005, 58, 97–105. [Google Scholar] [CrossRef] [PubMed]
- Whitten, M.; Sun, F.; Tew, I.; Soukou, C.; Nappi, A.; Ratcliffe, N. Differential modulation of Rhodnius prolixus nitric oxide activities following challenge with Trypanosoma rangeli, T. cruzi and bacterial cell wall components. Insect Biochem. Mol. Biol. 2007, 37, 440–452. [Google Scholar] [CrossRef] [PubMed]
- Hinnebusch, A.G. eIF3: A versatile scaffold for translation initiation complexes. Trends Biochem. Sci. 2006, 31, 553–562. [Google Scholar] [CrossRef]
- Graça-Souza, A.V.; Maya-Monteiro, C.; Paiva-Silva, G.O.; Braz, G.R.C.; Paes, M.C.; Sorgine, M.H.F.; Oliveira, M.F.; Oliveira, P.L. Adaptations against heme toxicity in blood-feeding arthropods. Insect Biochem. Mol. Biol. 2006, 36, 322–335. [Google Scholar] [CrossRef]
- Paiva-Silva, G.O.; Cruz-Oliveira, C.; Nakayasu, E.S.; Maya-Monteiro, C.M.; Dunkov, B.C.; Masuda, H.; Almeida, I.C.; Oliveira, P.L. A heme-degradation pathway in a blood-sucking insect. Proc. Natl. Acad. Sci. USA 2006, 103, 8030–8035. [Google Scholar] [CrossRef] [Green Version]
- Dansa-Petretski, M.; Ribeiro, J.M.C.; Atella, G.C.; Masuda, H.; Oliveira, P.L. Antioxidant role of Rhodnius prolixus heme-binding protein. Protection against heme-induced lipid peroxidation. J. Biol. Chem. 1995, 270, 10893–10896. [Google Scholar] [CrossRef] [Green Version]
- Oliveira, P.L.; Kawooya, J.K.; Ribeiro, J.M.C.; Meyer, T.; Poorman, R.; Alves, E.W.; Walker, F.A.; Machado, E.A.; Nussenzveig, R.H.; Padovan, G.J.; et al. A heme-binding protein from hemolymph and oocytes of the blood-sucking insect, Rhodnius prolixus. Isolation and characterization. J. Biol. Chem. 1995, 270, 10897–10901. [Google Scholar] [CrossRef] [Green Version]
- Oliveira, M.F.; Silva, J.R.; Dansa-Petretski, M.; De Souza, W.; Lins, U.; Braga, C.M.S.; Masuda, H.; Oliveira, P.L. Haem detoxification by an insect. Nature 1999, 400, 517–518. [Google Scholar] [CrossRef]
- Walter-Nuno, A.B.; Oliveira, M.P.; Oliveira, M.F.; Gonçalves, R.L.; Ramos, I.B.; Koerich, L.B.; Oliveira, P.L.; Paiva-Silva, G.O. Silencing of maternal heme-binding protein causes embryonic mitochondrial dysfunction and impairs embryogenesis in the blood sucking insect Rhodnius prolixus. J. Biol. Chem. 2013, 288, 29323–29332. [Google Scholar] [CrossRef] [Green Version]
- Taracena, M.L.; Oliveira, P.L.; Almendares, O.; Umaña, C.; Lowenberger, C.; Dotson, E.M.; Paiva-Silva, G.O.; Pennington, P.M. Genetically Modifying the Insect Gut Microbiota to Control Chagas Disease Vectors through Systemic RNAi. PLoS Negl. Trop. Dis. 2015, 9, e0003358. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Felton, G.W.; Summers, C.B. Antioxidant systems in insects. Arch. Insect Biochem. Physiol. 1995, 29, 187–197. [Google Scholar] [CrossRef] [PubMed]
- Yoshiga, T.; Hernandez, V.P.; Fallon, A.M.; Law, J.H. Mosquito transferrin, an acute-phase protein that is up-regulated upon infection. Proc. Natl. Acad. Sci. USA 1997, 94, 12337–12342. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gandara, A.C.P.; Oliveira, J.H.M.; Nunes, R.D.; Goncalves, R.L.S.; Dias, F.A.; Hecht, F.; Fernandes, D.C.; Genta, F.A.; Laurindo, F.R.M.; Oliveira, M.F.; et al. Amino acids trigger down-regulation of superoxide via TORC pathway in the midgut of Rhodnius prolixus. Biosci. Rep. 2016, 36, e00321. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Benoit, J.B.; Lopez-Martinez, G.; Patrick, K.R.; Phillips, Z.P.; Krause, T.B.; Denlinger, D.L. Drinking a hot blood meal elicits a protective heat shock response in mosquitoes. Proc. Natl. Acad. Sci. USA 2011, 108, 8026–8029. [Google Scholar] [CrossRef] [Green Version]
- Feder, M.E.; Hofmann, G.E. Heat-shock Proteins, Molecular Chaperones, and the Stress Response: Evolutionary and Ecological Physiology. Annu. Rev. Physiol. 1999, 61, 243–282. [Google Scholar] [CrossRef] [Green Version]
- Mahroof, R.; Kun, Y.Z.; Neven, L.; Subramanyam, B.; Bai, J. Expression patterns of three heat shock protein 70 genes among developmental stages of the red flour beetle, Tribolium castaneum (Coleoptera: Tenebrionidae). Comp. Biochem. Physiol. A Mol. Integr. Physiol. 2005, 141, 247–256. [Google Scholar] [CrossRef] [PubMed]
- Sanders, H.R.; Evans, A.M.; Ross, L.S.; Gill, S.S. Blood meal induces global changes in midgut gene expression in the disease vector, Aedes aegypti. Insect Biochem. Mol. Biol. 2003, 33, 1105–1122. [Google Scholar] [CrossRef]
- Kongsuwan, K.; Josh, P.; Zhu, Y.; Pearson, R.; Gough, J.; Colgrave, M.L. Exploring the midgut proteome of partially fed female cattle tick (Rhipicephalus (Boophilus) microplus). J. Insect Physiol. 2010, 56, 212–226. [Google Scholar] [CrossRef]
- Paim, R.M.M.; Araujo, R.N.; Leis, M.; Sant’anna, M.R.V.; Gontijo, N.F.; Lazzari, C.R.; Pereira, M.H. Functional evaluation of Heat Shock Proteins 70 (HSP70/HSC70) on Rhodnius prolixus (Hemiptera, Reduviidae) physiological responses associated with feeding and starvation. Insect Biochem. Mol. Biol. 2016, 77, 10–20. [Google Scholar] [CrossRef]
- Boulanger, N.; Bulet, P.; Lowenberger, C. Antimicrobial peptides in the interactions between insects and flagellate parasites. Trends Parasitol. 2006, 22, 262–268. [Google Scholar] [CrossRef]
- Flores-Villegas, A.L.; Salazar-Schettino, P.M.; Córdoba-Aguilar, A.; Gutiérrez-Cabrera, A.E.; Rojas-Wastavino, G.E.; Bucio-Torres, M.I.; Cabrera-Bravo, M. Immune defence mechanisms of triatomines against bacteria, viruses, fungi and parasites. Bull. Entomol. Res. 2015, 105, 523–532. [Google Scholar] [CrossRef]
- Kollien, A.H.; Fechner, S.; Waniek, P.J.; Schaub, G.A. Isolation and characterization of a cDNA encoding for a lysozyme from the gut of the reduviid bug Triatoma infestans. Arch. Insect Biochem. Physiol. 2003, 53, 134–145. [Google Scholar] [CrossRef]
- Ursic-Bedoya, R.J.; Nazzari, H.; Cooper, D.; Triana, O.; Wolff, M.; Lowenberger, C. Identification and characterization of two novel lysozymes from Rhodnius prolixus, a vector of Chagas disease. J. Insect Physiol. 2008, 54, 593–603. [Google Scholar] [CrossRef] [PubMed]
- Araujo, C.A.C.; Waniek, P.J.; Stock, P.; Mayer, C.; Jansen, A.M.; Schaub, G.A. Sequence characterization and expression patterns of defensin and lysozyme encoding genes from the gut of the reduviid bug Triatoma brasiliensis. Insect Biochem. Mol. Biol. 2006, 36, 547–560. [Google Scholar] [CrossRef] [PubMed]
- Inohara, N.; Nuez, G. ML—A conserved domain involved in innate immunity and lipid metabolism. Trends Biochem. Sci. 2002, 27, 219–221. [Google Scholar] [CrossRef]
- Kotsyfakis, M.; Schwarz, A.; Erhart, J.; Ribeiro, J.M.C. Tissue- and time-dependent transcription in Ixodes ricinus salivary glands and midguts when blood feeding on the vertebrate host. Sci. Rep. 2015, 5, 9103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paskewitz, S.M.; Shi, L. The hemolymph proteome of Anopheles gambiae. Insect Biochem. Mol. Biol. 2005, 35, 815–824. [Google Scholar] [CrossRef] [PubMed]
- Vieira, L.R.; Polomé, A.; Mesquita, R.D.; Salmon, D.; Braz, G.R.C.; Bousbata, S. Protein 2DE reference map of the anterior midgut of the blood-sucking bug Rhodnius prolixus. Proteomics 2015, 15, 3901–3904. [Google Scholar] [CrossRef] [PubMed]
- Gumiel, M.; De Mattos, D.P.; Vieira, C.S.; Gonzalez, M.S.; Teixeira-ferreira, A.; Waghabi, M. Proteome of the Triatomine Digestive Tract: From Catalytic to Immune Pathways; Focusing on Annexin Expression. Front. Mol. Biosci. 2020, 7, 589435. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ouali, R.; Vieira, L.R.; Salmon, D.; Bousbata, S. Early Post-Prandial Regulation of Protein Expression in the Midgut of Chagas Disease Vector Rhodnius prolixus Highlights New Potential Targets for Vector Control Strategy. Microorganisms 2021, 9, 804. https://doi.org/10.3390/microorganisms9040804
Ouali R, Vieira LR, Salmon D, Bousbata S. Early Post-Prandial Regulation of Protein Expression in the Midgut of Chagas Disease Vector Rhodnius prolixus Highlights New Potential Targets for Vector Control Strategy. Microorganisms. 2021; 9(4):804. https://doi.org/10.3390/microorganisms9040804
Chicago/Turabian StyleOuali, Radouane, Larissa Rezende Vieira, Didier Salmon, and Sabrina Bousbata. 2021. "Early Post-Prandial Regulation of Protein Expression in the Midgut of Chagas Disease Vector Rhodnius prolixus Highlights New Potential Targets for Vector Control Strategy" Microorganisms 9, no. 4: 804. https://doi.org/10.3390/microorganisms9040804






