CaptureSeq: Hybridization-Based Enrichment of cpn60 Gene Fragments Reveals the Community Structures of Synthetic and Natural Microbial Ecosystems
Abstract
:1. Introduction
2. Materials and Methods
2.1. CaptureSeq Array Design
2.2. Template DNA Preparation
2.3. Amplicon-Based Microbial Community Profiling
2.4. Whole Metagenome and CaptureSeq Sample Preparation
2.5. Reference Mapping
2.6. Sequence Assembly
2.7. Sub-OTU (sOTU) Definition Using Amplicon Sequence Variants (ASV)
2.8. Alpha Diversity Analysis
2.9. Beta Diversity Analysis
2.10. OTU Quantification
3. Results
3.1. CaptureSeq Provides Microbial Community Profiles from Synthetic Microbial Ecosystems
3.1.1. Zymobiomics Reference Panel
3.1.2. Quantification of Microbial Abundances in CaptureSeq
3.2. CaptureSeq Provides Microbial Community Profiles in Natural Ecosystems
3.3. CaptureSeq Data Facilitates the Assembly of Target OTU from Taxonomic Clusters
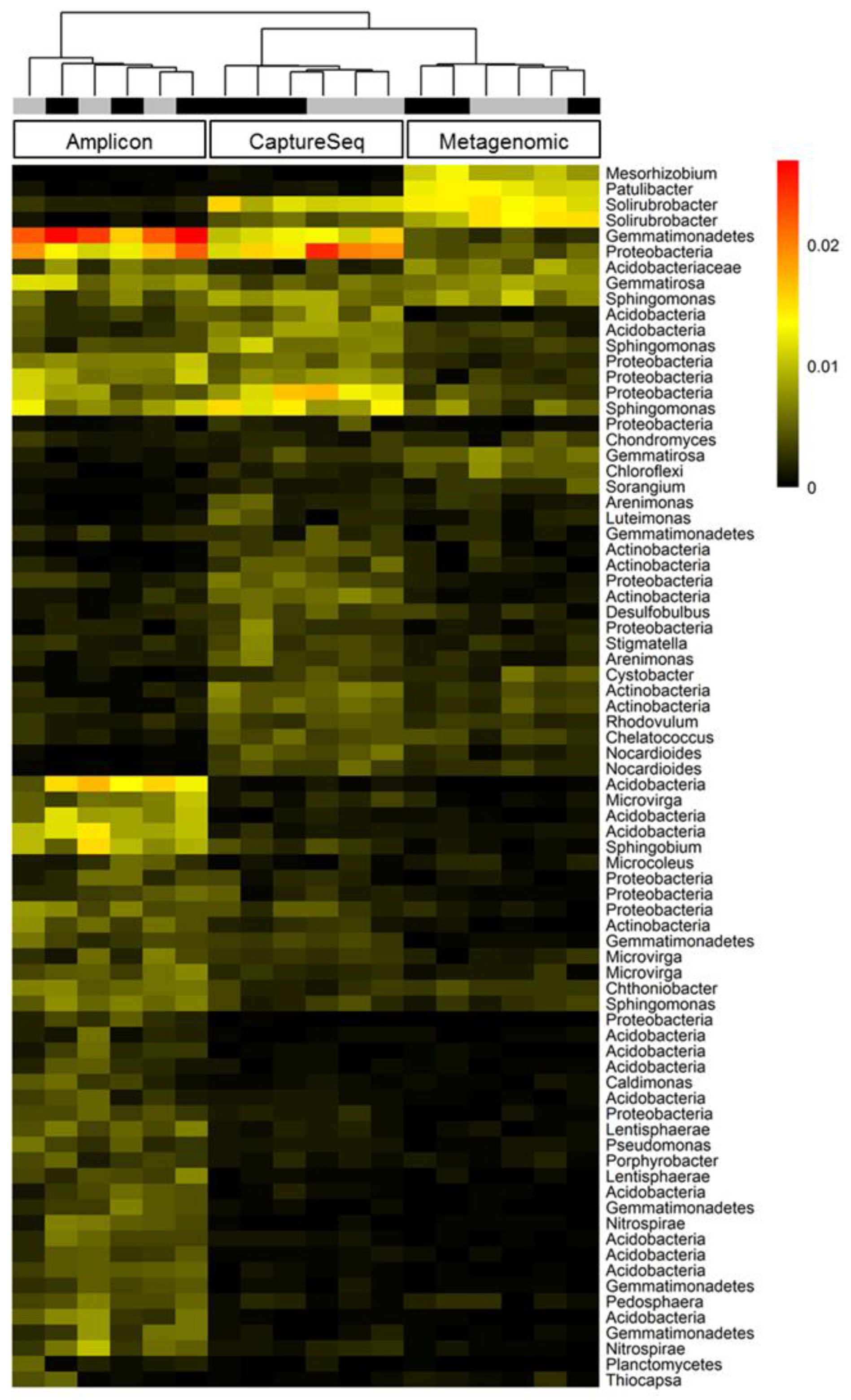
3.4. CaptureSeq Provides Sequencing Depth That Is Similar to That Achieved by Taxonomic Marker PCR Amplification
3.5. Microbial Ecosystem Diversity Metrics of Antibiotic-Treated Soil Samples
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Woese, C.R.; Fox, G.E. Phylogenetic structure of the prokaryotic domain: The primary kingdoms. Proc. Natl. Acad. Sci. USA 1977, 74, 5088–5090. [Google Scholar] [CrossRef] [Green Version]
- Woese, C.R.; Kandler, O.; Wheelis, M.L. Towards a natural system of organisms: Proposal for the domains Archaea, Bacteria, and Eucarya. Proc. Natl. Acad. Sci. USA 1990, 87, 4576–4579. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tikhonovich, I.; Provorov, N. Microbiology is the basis of sustainable agriculture: An opinion. Ann. Appl. Biol. 2011, 159, 155–168. [Google Scholar] [CrossRef]
- Weller, R.; Ward, D.M. Selective recovery of 16S rRNA sequences from natural microbial communities in the form of cDNA. Appl. Environ. Microbiol. 1989, 55, 1818–1822. [Google Scholar] [CrossRef] [Green Version]
- Staley, J.T.; Konopka, A. Measurement of in situ activities of nonphotosynthetic microorganisms in aquatic and terrestrial habitats. Appl. Environ. Microbiol. 1985, 39, 321–346. [Google Scholar] [CrossRef] [PubMed]
- Edge, T.A.; Baird, D.J.; Bilodeau, G.; Gagné, N.; Greer, C.; Konkin, D.; Newton, G.; Séguin, A.; Beaudette, L.; Bilkhu, S.; et al. The Ecobiomics project: Advancing metagenomics assessment of soil health and freshwater quality in Canada. Sci. Total Environ. 2020, 710, 135906. [Google Scholar] [CrossRef]
- Turnbaugh, P.J.; Ley, R.E.; Hamady, M.; Fraser-Liggett, C.M.; Knight, R.; Gordon, J.I. The Human Microbiome Project. Nature 2007, 449, 804–810. [Google Scholar] [CrossRef]
- Hebert, P.D.N.; Cywinska, A.; Ball, S.L.; Dewaard, J.R. Biological identifications through DNA barcodes. Proc. R. Soc. Lond. Ser. B Biol. Sci. 2003, 270, 313–321. [Google Scholar] [CrossRef] [Green Version]
- Singer, E.; Bushnell, B.; Coleman-Derr, D.; Bowman, B.; Bowers, R.M.; Levy, A.; A Gies, E.; Cheng, J.-F.; Copeland, A.; Klenk, H.-P.; et al. High-resolution phylogenetic microbial community profiling. ISME J. 2016, 10, 2020–2032. [Google Scholar] [CrossRef]
- Hill, J.E.; Penny, S.L.; Crowell, K.G.; Goh, S.H.; Hemmingsen, S.M. cpnDB: A chaperonin sequence database. Genome Res. 2004, 14, 1669–1675. [Google Scholar] [CrossRef] [Green Version]
- Adékambi, T.; Drancourt, M.; Raoult, D. The rpoB gene as a tool for clinical microbiologists. Trends Microbiol. 2009, 17, 37–45. [Google Scholar] [CrossRef]
- Barret, M.; Gagnon, N.; Kalmokoff, M.L.; Topp, E.; Verastegui, Y.; Brooks, S.P.J.; Matias, F.; Neufeld, J.D.; Talbot, G. Identification of Methanoculleus spp. as active methanogens during anoxic incubations of swine manure storage tank samples. Appl. Environ. Microbiol. 2013, 79, 424–433. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schoch, C.L.; Seifert, K.A.; Huhndorf, S.; Robert, V.; Spouge, J.L.; Levesque, C.A.; Chen, W.; Barcoding, C.F. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc. Natl. Acad. Sci. USA 2012, 109, 6241–6246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barret, M.; Briand, M.; Bonneau, S.; Préveaux, A.; Valière, S.; Bouchez, O.; Hunault, G.; Simoneau, P.; Jacques, M.-A. Emergence shapes the structure of the seed-microbiota. Appl. Environ. Microbiol. 2015, 81, 1257–1266. [Google Scholar] [CrossRef] [Green Version]
- Walker, A.W.; Martin, J.C.; Scott, P.T.; Parkhill, J.; Flint, H.J.; Scott, K.P. 16S rRNA gene-based profiling of the human infant gut microbiota is strongly influenced by sample processing and PCR primer choice. Microbiome 2015, 3, 26. [Google Scholar] [CrossRef] [Green Version]
- Guo, J.; Cole, J.R.; Zhang, Q.; Brown, C.T.; Tiedje, J.M. Microbial community analysis with ribosomal gene fragments from shotgun metagenomes. Appl. Environ. Microbiol. 2016, 82, 157–166. [Google Scholar] [CrossRef] [Green Version]
- Lynch, M.D.J.; Neufeld, J.D. Ecology and exploration of the rare biosphere. Nat. Rev. Microbiol. 2015, 13, 217–229. [Google Scholar] [CrossRef]
- Poretsky, R.; Rodriguez-R, L.M.; Luo, C.; Tsementzi, D.; Konstantinidis, K.T. Strengths and limitations of 16S rRNA gene amplicon sequencing in revealing temporal microbial community dynamics. PLoS ONE 2014, 9, e93827. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hess, M.; Sczyrba, A.; Egan, R.; Kim, T.-W.; Chokhawala, H.; Schroth, G.; Luo, S.; Clark, D.S.; Chen, F.; Zhang, T.; et al. Metagenomic discovery of biomass-degrading genes and genomes from cow rumen. Science 2011, 331, 463–467. [Google Scholar] [CrossRef] [Green Version]
- Raymond, F.; Ouameur, A.A.; Deraspe, M.; Iqbal, N.; Gingras, H.; Dridi, B.; Leprohon, P.; Plante, P.-L.; Giroux, R.; Berube, E.; et al. The initial state of the human gut microbiome determines its reshaping by antibiotics. ISME J. 2016, 10, 707–720. [Google Scholar] [CrossRef] [PubMed]
- Handley, K.M.; VerBerkmoes, N.C.; I Steefel, C.; Williams, K.H.; Sharon, I.; Miller, C.S.; Frischkorn, K.R.; Chourey, K.; Thomas, B.C.; Shah, M.B.; et al. Biostimulation induces syntrophic interactions that impact C, S and N cycling in a sediment microbial community. ISME J. 2013, 7, 800–816. [Google Scholar] [CrossRef] [Green Version]
- Fierer, N.; Leff, J.W.; Adams, B.J.; Nielsen, U.N.; Bates, S.T.; Lauber, C.L.; Owens, S.; Gilbert, J.A.; Wall, D.H.; Caporaso, J.G. Cross-biome metagenomic analyses of soil microbial communities and their functional attributes. Proc. Natl. Acad. Sci. USA 2012, 109, 21390–21395. [Google Scholar] [CrossRef] [Green Version]
- Luo, C.; Rodriguez-R, L.M.; Johnston, E.R.; Wu, L.; Cheng, L.; Xue, K.; Tu, Q.; Deng, Y.; He, Z.; Shi, J.Z.; et al. Soil microbial community responses to a decade of warming as revealed by comparative metagenomics. Appl. Environ. Microbiol. 2014, 80, 1777–1786. [Google Scholar] [CrossRef] [Green Version]
- Chaban, B.; E Hill, J. A ’universal’ type II chaperonin PCR detection system for the investigation of Archaea in complex microbial communities. ISME J. 2012, 6, 430–439. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Links, M.G.; Demeke, T.; Gräfenhan, T.; Hill, J.E.; Hemmingsen, S.M.; Dumonceaux, T.J. Simultaneous profiling of seed-associated bacteria and fungi reveals antagonistic interactions between microorganisms within a shared epiphytic microbiome on Triticum and Brassica seeds. New Phytol. 2014, 202, 542–553. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Verbeke, T.J.; Sparling, R.; Hill, J.E.; Links, M.G.; Levin, D.; Dumonceaux, T.J. Predicting relatedness of bacterial genomes using the chaperonin-60 universal target (cpn60 UT): Application to Thermoanaerobacter species. Syst. Appl. Microbiol. 2011, 34, 171–179. [Google Scholar] [CrossRef] [PubMed]
- Jayaprakash, T.P.; Schellenberg, J.J.; Hill, J.E. Resolution and Characterization of Distinct cpn60-Based Subgroups of Gardnerella vaginalis in the Vaginal Microbiota. PLoS ONE 2012, 7, e43009. [Google Scholar] [CrossRef]
- Tian, Q.; Zhao, W.; Lu, S.; Zhu, S.; Li, S. DNA Barcoding for Efficient Species- and Pathovar-Level Identification of the Quarantine Plant Pathogen Xanthomonas. PLoS ONE 2016, 11, e0165995. [Google Scholar] [CrossRef]
- Links, M.G.; Dumonceaux, T.J.; Hemmingsen, S.M.; Hill, J.E. The chaperonin-60 universal target is a barcode for bacteria that enables de novo assembly of metagenomic sequence data. PLoS ONE 2012, 7, e49755. [Google Scholar] [CrossRef] [Green Version]
- Links, M.G.; Chaban, B.; Hemmingsen, S.M.; Muirhead, K.; E Hill, J. mPUMA: A computational approach to microbiota analysis by de novo assembly of operational taxonomic units based on protein-coding barcode sequences. Microbiome 2013, 1, 23. [Google Scholar] [CrossRef] [Green Version]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef] [Green Version]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581. [Google Scholar] [CrossRef] [Green Version]
- Knight, R.; Vrbanac, A.; Taylor, B.C.; Aksenov, A.; Callewaert, C.; Debelius, J.; Gonzalez, A.; Kosciolek, T.; McCall, L.-I.; McDonald, D.; et al. Best practices for analysing microbiomes. Nat. Rev. Microbiol. 2018, 16, 410–422. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vancuren, S.J.; Dos Santos, S.J.; Hill, J.E. Maternal Microbiome Legacy Project, Evaluation of variant calling for cpn60 barcode sequence-based microbiome profiling. PLoS ONE 2020, 15, e0235682. [Google Scholar] [CrossRef]
- Hill, J.E.; Town, J.R.; Hemmingsen, S.M. Improved template representation in cpn60 polymerase chain reaction (PCR) product libraries generated from complex templates by application of a specific mixture of PCR primers. Environ. Microbiol. 2006, 8, 741–746. [Google Scholar] [CrossRef]
- Dumonceaux, T.J.; Hill, J.E.; Hemmingsen, S.M.; van Kessel, A.G. Characterization of intestinal microbiota and response to dietary virginiamycin supplementation in the broiler chicken. Appl. Environ. Microbiol. 2006, 72, 2815–2823. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bos, K.I.; Herbig, A.; Sahl, J.; Waglechner, N.; Fourment, M.; Forrest, S.A.; Klunk, J.; Schuenemann, V.J.; Poinar, D.; Kuch, M.; et al. Eighteenth century Yersinia pestis genomes reveal the long-term persistence of an historical plague focus. ELife 2016, 5, e12994. [Google Scholar] [CrossRef] [PubMed]
- Seifert, L.; Wiechmann, I.; Harbeck, M.; Thomas, A.; Grupe, G.; Projahn, M.; Scholz, H.C.; Riehm, J.M. Genotyping Yersinia pestis in historical plague: Evidence for long-term persistence of Y.pestis in Europe from the 14th to the 17th century. PLoS ONE 2016, 11, e0145194. [Google Scholar] [CrossRef] [Green Version]
- Wagner, D.M.; Klunk, J.; Harbeck, M.; Devault, A.; Waglechner, N.; Sahl, J.W.; Enk, J.; Birdsell, D.N.; Kuch, M.; Lumibao, C.; et al. Yersinia pestis and the Plague of Justinian 541-543 AD: A genomic analysis. Lancet Infect. Dis. 2014, 14, 319–326. [Google Scholar] [CrossRef]
- Gasc, C.; Peyret, P. Hybridization capture reveals microbial diversity missed using current profiling methods. Microbiome 2018, 6, 61. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hill, J.E.; Vancuren, S.J. Update on cpnDB: A reference database of chaperonin sequences. Database 2019, 2019. [Google Scholar] [CrossRef]
- Dumonceaux, T.; Links, M.; Town, J.; Hill, J.; Hemmingsen, S. Targeted capture of cpn60 gene fragments for PCR-independent microbial community profiling. Nat. Protoc. Exch. 2017. [Google Scholar] [CrossRef]
- Cleary, D.W.; Bishop, A.H.; Zhang, L.; Topp, E.; Wellington, E.M.; Gaze, W.H. Long-term antibiotic exposure in soil is associated with changes in microbial community structure and prevalence of class 1 integrons. FEMS Microbiol. Ecol. 2016, 92, fiw159. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caporaso, J.G.; Ackermann, G.; Apprill, A.; Bauer, M.; Berg-Lyons, D.; Betley, J.; Fierer, N.; Fraser, L.; Fuhrman, J.A.; Gilbert, J.A.; et al. EMP 16S Illumina Amplicon Protocol. Protcols.io. 2018. Available online: https://www.protocols.io/view/emp-16s-illumina-amplicon-protocol-nuudeww (accessed on 1 April 2021).
- Hill, J.; Fernando, C. cpn60 metagenomic amplicon library preparation for the Illumina Miseq platform. Nat. Protoc. Exch. 2021. [Google Scholar] [CrossRef]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009, 10, R25. [Google Scholar] [CrossRef] [Green Version]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. Embnet J. 2011, 17, 3. [Google Scholar] [CrossRef]
- Magoc, T.; Salzberg, S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [Green Version]
- Rozen, S.; Skaletsky, H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 2000, 132, 365–386. [Google Scholar] [PubMed] [Green Version]
- Pérez-López, E.; Hammond, C.; Olivier, C.Y.; Dumonceaux, T.J. Detection and Typing of ‘Candidatus Phytoplasma’ spp. in Host DNA extracts Using Oligonucleotide-Coupled Fluorescent Microspheres. In Diagnostic Bacteriology; Bishop-Lilly, K.A., Ed.; Humana Press: New York, NY, USA, 2017. [Google Scholar]
- Dumonceaux, T.J.; Schellenberg, J.; Goleski, V.; Hill, J.E.; Jaoko, W.; Kimani, J.; Money, D.; Ball, T.B.; Plummer, F.A.; Severini, A. Multiplex detection of bacteria associated with normal microbiota and with bacterial vaginosis in vaginal swabs by use of oligonucleotide-coupled fluorescent microspheres. J. Clin. Microbiol. 2009, 47, 4067–4077. [Google Scholar] [CrossRef] [Green Version]
- Lee, D.H.; Zo, Y.G.; Kim, S.J. Nonradioactive method to study genetic profiles of natural bacterial communities by PCR-single-strand-conformation polymorphism. Appl. Environ. Microbiol. 1996, 62, 3112–3120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Topp, E.; Chapman, R.; Devers-Lamrani, M.; Hartmann, A.; Marti, R.; Martin-Laurent, F.; Sabourin, L.; Scott, A.; Sumarah, M. Accelerated biodegradation of veterinary antibiotics in agricultural soil following long-term exposure, and isolation of a sulfamethazine-degrading Microbacterium sp. J. Environ. Qual. 2013, 42, 173–178. [Google Scholar] [CrossRef]
- Martin-Laurent, F.; Marti, R.; Waglechner, N.; Wright, G.D.; Topp, E. Draft genome sequence of the sulfonamide antibiotic-degrading Microbacterium sp. strain C448. Genome Announc. 2014, 2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chao, A.; Hsieh, T.C.; Chazdon, R.L.; Colwell, R.K.; Gotelli, N.J. Unveiling the species-rank abundance distribution by generalizing the Good-Turing sample coverage theory. Ecology 2015, 96, 1189–1201. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Edgar, R.C. Accuracy of microbial community diversity estimated by closed- and open-reference OTUs. PeerJ 2017, 5, e3889. [Google Scholar] [CrossRef] [Green Version]
- Ranjan, R.; Rani, A.; Metwally, A.; McGee, H.S.; Perkins, D.L. Analysis of the microbiome: Advantages of whole genome shotgun versus 16S amplicon sequencing. Biochem. Biophys. Res. Commun. 2016, 469, 967–977. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schwartz, M.H.; Wang, H.; Pan, J.N.; Clark, W.C.; Cui, S.; Eckwahl, M.J.; Pan, D.W.; Parisien, M.; Owens, S.M.; Cheng, B.L.; et al. Microbiome characterization by high-throughput transfer RNA sequencing and modification analysis. Nat. Commun. 2018, 9, 5353. [Google Scholar] [CrossRef]
- McKenney, E.A.; Ashwell, M.; Lambert, J.E.; Fellner, V. Fecal microbial diversity and putative function in captive western lowland gorillas (Gorilla gorilla gorilla), common chimpanzees (Pan troglodytes), Hamadryas baboons (Papio hamadryas) and binturongs (Arctictis binturong). Integr. Zool. 2014, 9, 557–569. [Google Scholar] [CrossRef] [PubMed]
- Town, J.R.; Links, M.G.; Fonstad, T.A.; Dumonceaux, T.J. Molecular characterization of anaerobic digester microbial communities identifies microorganisms that correlate to reactor performance. Bioresour. Technol. 2014, 151, 249–257. [Google Scholar] [CrossRef]
- Peterson, S.W.; Knox, N.C.; Golding, G.R.; Tyler, S.D.; Tyler, A.D.; Mabon, P.; Embree, J.E.; Fleming, F.; Fanella, S.; van Domselaar, G.; et al. A Study of the Infant Nasal Microbiome Development over the First Year of Life and in Relation to Their Primary Adult Caregivers Using cpn60 Universal Target (UT) as a Phylogenetic Marker. PLoS ONE 2016, 11, e0152493. [Google Scholar] [CrossRef] [Green Version]
- Hill, J.E.; Hemmingsen, S.M.; Goldade, B.G.; Dumonceaux, T.J.; Klassen, J.; Zijlstra, R.T.; Goh, S.H.; van Kessel, A.G. Comparison of ileum microflora of pigs fed corn-, wheat-, or barley-based diets by chaperonin-60 sequencing and quantitative PCR. Appl. Environ. Microbiol. 2005, 71, 867–875. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robertson, G.; Schein, J.; Chiu, R.; Corbett, R.; Field, M.; Jackman, S.D.; Mungall, K.; Lee, S.; Okada, H.M.; Qian, J.Q.; et al. De novo assembly and analysis of RNA-seq data. Nat. Methods 2010, 7, 909–912. [Google Scholar] [CrossRef] [PubMed]
- Cooper, R.M.; Tsimring, L.; Hasty, J. Inter-species population dynamics enhance microbial horizontal gene transfer and spread of antibiotic resistance. ELife 2017, 6, e25950. [Google Scholar] [CrossRef]
- Props, R.; Kerckhof, F.-M.; Rubbens, P.; de Vrieze, J.; Sanabria, E.H.; Waegeman, W.; Monsieurs, P.; Hammes, F.; Boon, N. Absolute quantification of microbial taxon abundances. ISME J. 2016, 11, 584–587. [Google Scholar] [CrossRef] [Green Version]
- Johnson, L.A.; Chaban, B.; Harding, J.C.; Hill, J.E. Optimizing a PCR protocol for cpn60-based microbiome profiling of samples variously contaminated with host genomic DNA. BMC Res. Notes 2015, 8, 253. [Google Scholar] [CrossRef] [Green Version]
- Green, S.J.; Venkatramanan, R.; Naqib, A. Deconstructing the polymerase chain reaction: Understanding and correcting bias associated with primer degeneracies and primer-template mismatches. PLoS ONE 2015, 10, e0128122. [Google Scholar] [CrossRef]
- Lee, C.K.; Herbold, C.W.; Polson, S.W.; Wommack, K.E.; Williamson, S.J.; McDonald, I.R.; Cary, S.C. Groundtruthing next-gen sequencing for microbial ecology-biases and errors in community structure estimates from PCR amplicon pyrosequencing. PLoS ONE 2012, 7, e44224. [Google Scholar] [CrossRef] [Green Version]
- Pinto, A.J.; Raskin, L. PCR biases distort bacterial and archaeal community structure in pyrosequencing datasets. PLoS ONE 2012, 7, e43093. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Demirel, B.; Scherer, P. The roles of acetotrophic and hydrogenotrophic methanogens during anaerobic conversion of biomass to methane: A review. Rev. Environ. Sci. Biotechnol. 2008, 7, 173–190. [Google Scholar] [CrossRef]
- de Menezes, A.B.; Richardson, A.E.; Thrall, P.H. Linking fungal–bacterial co-occurrences to soil ecosystem function. Curr. Opin. Microbiol. 2017, 37, 135–141. [Google Scholar] [CrossRef]
- Carballa, M.; Regueiro, L.; Lema, J.M. Microbial management of anaerobic digestion: Exploiting the microbiome-functionality nexus. Curr. Opin. Microbiol. 2015, 33, 103–111. [Google Scholar] [CrossRef]
- Gopal, M.; Gupta, A. Microbiome selection could spur next-generation plant breeding strategies. Front. Microbiol. 2016, 7, 1971. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muegge, B.D.; Kuczynski, J.; Knights, D.; Clemente, J.C.; González, A.; Fontana, L.; Henrissat, B.; Knight, R.; Gordon, J.I. Diet drives convergence in gut microbiome functions across mammalian phylogeny and within humans. Science 2011, 332, 970–974. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kau, A.L.; Ahern, P.P.; Griffin, N.W.; Goodman, A.L.; Gordon, J.I. Human nutrition, the gut microbiome and the immune system. Nature 2011, 474, 327–336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Busby, P.E.; Soman, C.; Wagner, M.R.; Friesen, M.L.; Kremer, J.; Bennett, A.; Morsy, M.; Eisen, J.A.; Leach, J.E.; Dangl, J.L. Research priorities for harnessing plant microbiomes in sustainable agriculture. PLoS Biol. 2017, 15, e2001793. [Google Scholar] [CrossRef]
- Koch, C.; Müller, S.; Harms, H.; Harnisch, F. Microbiomes in bioenergy production: From analysis to management. Curr. Opin. Biotechnol. 2014, 27, 65–72. [Google Scholar] [CrossRef] [PubMed]
- Fierer, N. Embracing the unknown: Disentangling the complexities of the soil microbiome. Nat. Rev. Microbiol. 2017, 15, 579–590. [Google Scholar] [CrossRef] [PubMed]






| CaptureSeq Assembly 1 | Shotgun Assembly 2 | sOTU Detected 3 | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Organism | cpn60 UT Sequence Length | OTU Detected | Assembly Length (bp) | Sequence Identity 4 | cpn60 OTU Detected | Assembly Length (bp) | Sequence Identity | cpn60 CaptureSeq | 16S Amplification |
| Prokaryotes | |||||||||
| Bacillus subtilis | 552 | + | 2144 | 100% | + | 873 | 100% | + | + |
| Escherichia coli | 555 | + | 1138 | 97% | + | 1208 | 99% | + | + |
| Enterococcus faecalis | 552 | + | 1235 | 100% | + | 1133 | 100% | + | + |
| Lactobacillus fermentum | 552 | + | 2165 | 100% | + | 870 | 100% | + | + |
| Listeria monocytogenes | 552 | + | 1589 | 100% | + | 811 | 99% | + | + |
| Pseudomonas aeruginosa | 555 | + | 1183 | 100% | + | 668 | 93% | + | + |
| Staphylococcus aureus | 552 | + | 1849 | 100% | + | 487 | 99% | + | + |
| Salmonella enterica | 555 | + | 1138 | 97% | + | 1366 | 100% | + | + |
| Eukaryotes | |||||||||
| Saccharomyces cerevisiae | 555 | + | 1301 | 100% | + | 384 | 94% | NF 5 | NF |
| Cryptococcus neoformans | 619 6 | + | 1435 | 100% | + | 549 | 94% | + | NF |
| Spike Level | High | Medium | Low | Unspiked | ||||
|---|---|---|---|---|---|---|---|---|
| Microorganism | Pre-Hyb | Post-Hyb | Pre-Hyb | Post-Hyb | Pre-Hyb | Post-Hyb | Pre-Hyb | Post-Hyb |
| G. vaginalis1 | 7.18 (0.85) | 7.58 (1.36) | 6.25 (0.72) | 6.87 (1.17) | 5.22 (0.61) | 5.54 (0.99) | 0.00 (0.00) | 0.00 (0.00) |
| L. crispatus1 | 7.20 (0.85) | 7.49 (1.35) | 6.26 (0.72) | 6.81 (1.16) | 5.28 (0.61) | 5.45 (0.98) | 0.00 (0.00) | 0.00 (0.00) |
| L. gasseri1 | 7.36 (0.87) | 8.29 (1.49) | 6.34 (0.73) | 7.56 (1.28) | 5.37 (0.62) | 6.33 (1.14) | 0.00 (0.00) | 0.00 (0.00) |
| A. vaginae1 | 7.24 (0.86) | 7.46 (1.34) | 6.36 (0.73) | 6.77 (1.15) | 5.39 (0.63) | 5.49 (0.99) | 0.00 (0.00) | 0.00 (0.00) |
| L. iners1 | 7.11 (0.84) | 7.42 (1.33) | 6.21 (0.72) | 6.59 (1.12) | 5.28 (0.61) | 5.45 (0.98) | 0.00 (0.00) | 0.00 (0.00) |
| P. agglomerans2 | 4.98 (0.59) | 6.43 (1.16) | 5.09 (0.59) | 6.78 (1.15) | 5.04 (0.58) | 6.44 (1.16) | 5.20 (0.61) | 6.51 (1.13) |
| Alternaria sp. 2 | 5.36 (0.63) | 4.76 (0.86) | 5.55 (0.64) | 5.19 (0.88) | 5.55 (0.64) | 4.96 (0.89) | 5.61 (0.65) | 5.12 (0.89) |
| 16S 2 | 8.46 | 5.56 | 8.67 | 5.89 | 8.62 | 5.56 | 8.59 | 5.74 |
| Pearson (r2) | |||
|---|---|---|---|
| Organism | CaptureSeq | Amplification | n |
| Gardnerella vaginalis | 0.999 | 0.724 | 4 |
| Lactobacillus iners | 0.998 | 0.926 | 4 |
| Lactobacillus crispatus | 0.998 | 0.922 | 4 |
| Lactobacillus gasseri | 1.000 | 0.937 | 4 |
| Atopobium vaginae | 1.000 | 0.782 | 4 |
| Spearman (ρ) | |||
| All combined | 0.956 | 0.912 | 20 |
| OTU | Domain | cpnDB Nearest Neighbor | OTU Length (bp) | Sequence Identity (%) 1 | Treatment (mg kg−1) | Soil Extract (Copies/g Soil) | Post-Hybridization Sample (Copies/µL) |
|---|---|---|---|---|---|---|---|
| XP002901426 DN2_c0_g1_i1 | Eukarya (type I) 2 | Phytophthora infestans | 539 | 100 | 0 | ND 3 | 1242 |
| 10 | ND | 3942 | |||||
| WP036300323 DN4_c3_g1_i2 | Bacteria (type I) | Microbacterium lacus C448 | 1066 | 99 | 0 | 6750 | 1417 |
| 10 | 38,571 4 | 8170 4 | |||||
| KUL05486 DN0_c0_g1_i1 | Archaea (type II) | Methanoculleus marisnigri | 1029 | 92 | 0 | 495 | ND |
| 10 | 527 | 3360 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Links, M.G.; Dumonceaux, T.J.; McCarthy, E.L.; Hemmingsen, S.M.; Topp, E.; Town, J.R. CaptureSeq: Hybridization-Based Enrichment of cpn60 Gene Fragments Reveals the Community Structures of Synthetic and Natural Microbial Ecosystems. Microorganisms 2021, 9, 816. https://doi.org/10.3390/microorganisms9040816
Links MG, Dumonceaux TJ, McCarthy EL, Hemmingsen SM, Topp E, Town JR. CaptureSeq: Hybridization-Based Enrichment of cpn60 Gene Fragments Reveals the Community Structures of Synthetic and Natural Microbial Ecosystems. Microorganisms. 2021; 9(4):816. https://doi.org/10.3390/microorganisms9040816
Chicago/Turabian StyleLinks, Matthew G., Tim J. Dumonceaux, E. Luke McCarthy, Sean M. Hemmingsen, Edward Topp, and Jennifer R. Town. 2021. "CaptureSeq: Hybridization-Based Enrichment of cpn60 Gene Fragments Reveals the Community Structures of Synthetic and Natural Microbial Ecosystems" Microorganisms 9, no. 4: 816. https://doi.org/10.3390/microorganisms9040816
APA StyleLinks, M. G., Dumonceaux, T. J., McCarthy, E. L., Hemmingsen, S. M., Topp, E., & Town, J. R. (2021). CaptureSeq: Hybridization-Based Enrichment of cpn60 Gene Fragments Reveals the Community Structures of Synthetic and Natural Microbial Ecosystems. Microorganisms, 9(4), 816. https://doi.org/10.3390/microorganisms9040816








