Moonlighting in Bacillus subtilis: The Small Proteins SR1P and SR7P Regulate the Moonlighting Activity of Glyceraldehyde 3-Phosphate Dehydrogenase A (GapA) and Enolase in RNA Degradation
Abstract
1. Introduction
2. Moonlighting Proteins in Bacillus Subtilis Known until 2015
2.1. TilS-HprT—A Complex of Two Enzymes—Acts as Transcriptional Activator at the ftsH Promoter
2.2. The Aconitase CitB Moonlights as RNA Binding Protein
2.3. The Glucose Permease PtsG Phosphorylates the RNA Binding Protein GlcT
2.4. The Glutamine Synthetase GlnA Interacts with the Transcription Factor TnrA and Inactivates Its DNA Binding Activity
2.5. Other Moonlighting Proteins
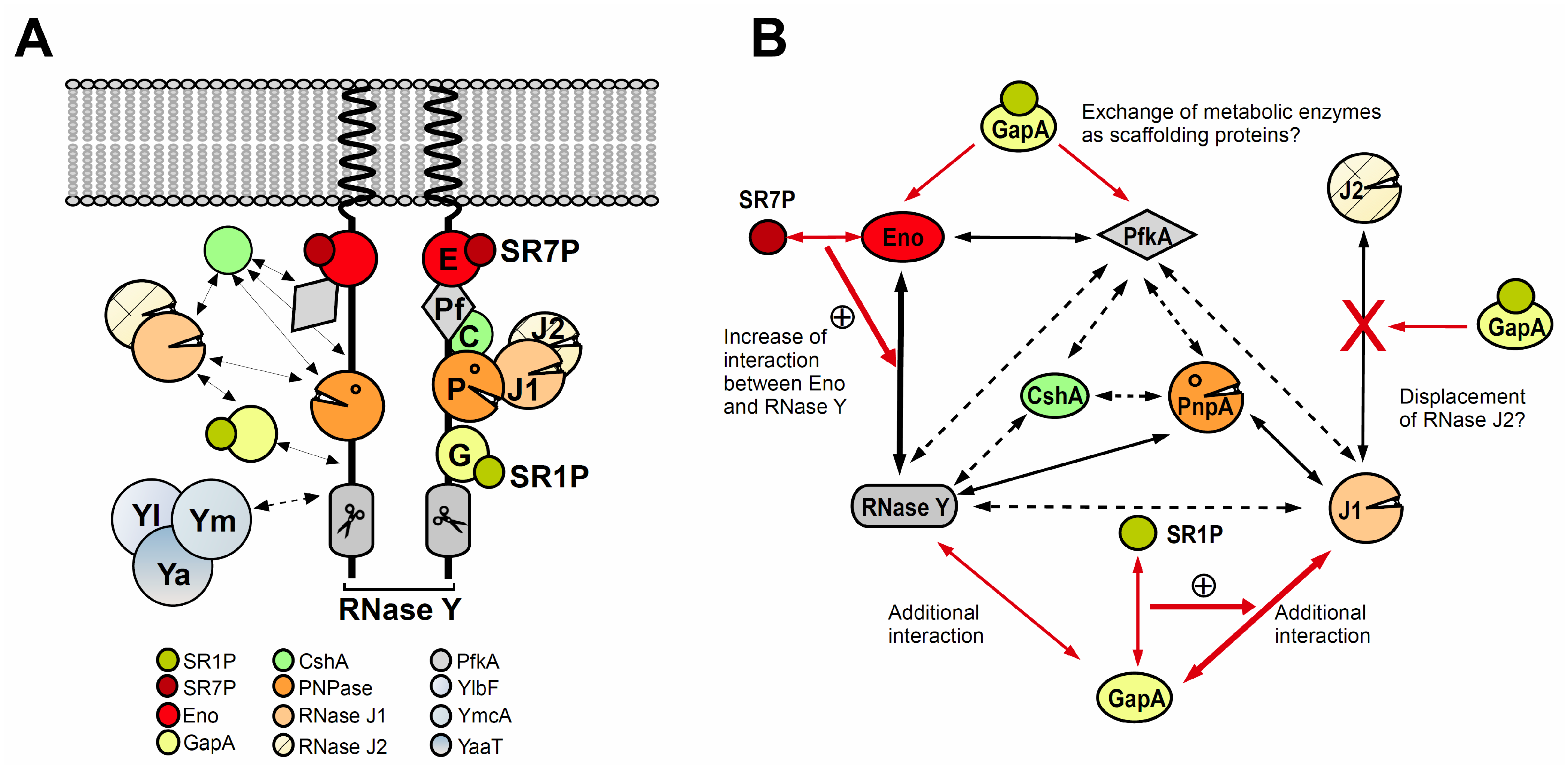
3. The Bacillus Subtilis Degradosome-Like Network (DLN)
Composition of the B. Subtilis DLN
4. GapA–SR1P
4.1. GapA, the Glycolytic Glyceraldehyde-3P-Dehydrogenase in Bacillus Subtilis, Can Bind Two RNases, RNase J1 and RNase Y and Moonlights in RNA Degradation
4.2. The Small Protein SR1P Impacts the Moonlighting Activity of GapA
5. Eno–SR7P
5.1. Enolase Moonlighting in B. Subtilis and Other Bacteria
5.2. The Small Protein SR7P Modulates the Moonlighting Activity of Enolase
6. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Henderson, B.; Martin, A. Bacterial virulence in the moonlight: Multitasking bacterial moonlighting proteins are virulence determinants in infections disease. Infect. Immun. 2011, 79, 3476–3491. [Google Scholar] [CrossRef]
- Pancholi, V.; Fischetti, V.A. A major surface protein on group A streptococci is a glyceraldehyde-3-phosphate-dehydrogenase with multiple binding activity. J. Exp. Med. 1992, 176, 415–426. [Google Scholar] [CrossRef]
- Winram, S.B.; Lottenberg, R. The plasmin-binding protein Plr of group A streptococci is identified as glyceraldehyde-3-phosphate dehydrogenase. Microbiology 1996, 142, 2311–2320. [Google Scholar] [CrossRef]
- Commichau, F.M.; Stülke, J. Trigger enzymes: Bifunctional proteins active in metabolism and in controlling gene expression. Mol. Microbiol. 2008, 67, 692–702. [Google Scholar] [CrossRef]
- Commichau, F.M.; Stülke, J. Trigger enzymes: Coordination of metabolism and virulence gene expression. Microbiol. Spectr. 2015, 3. [Google Scholar] [CrossRef]
- Commichau, F.M.; Rothe, F.M.; Herzberg, C.; Wagner, E.; Hellwig, D.; Lehnik-Habrink, M.; Hammer, E.; Völker, U.; Stülke, J. Novel activities of glycolytic enzymes in Bacillus subtilis: Interactions with essential proteins involved in mRNA processing. Mol. Cell. Proteom. 2009, 8, 1350–1360. [Google Scholar] [CrossRef]
- Licht, A.; Preis, S.; Brantl, S. Implication of CcpN in the regulation of a novel untranslated RNA (SR1) in Bacillus subtilis. Mol. Microbiol. 2005, 58, 189–206. [Google Scholar] [CrossRef] [PubMed]
- Gimpel, M.; Heidrich, N.; Mäder, U.; Krügel, H.; Brantl, S. A dual-function sRNA from B. subtilis: SR1 acts as a peptide encoding mRNA on the gapA operon. Mol. Microbiol. 2010, 76, 880–1009. [Google Scholar] [CrossRef]
- Gimpel, M.; Brantl, S. Dual-function sRNA encoded peptide SR1P modulates moonlighting activity of B. subtilis GapA. RNA Biol. 2016, 13, 916–926. [Google Scholar] [CrossRef]
- Gimpel, M.; Brantl, S. Dual-function small regulatory RNAs in bacteria. Mol. Microbiol. 2017, 103, 387–397. [Google Scholar] [CrossRef]
- Ul Haq, I.; Müller, P.; Brantl, S. SR7—A dual-function antisense RNA from Bacillus subtilis. RNA Biol. 2021, 18, 104–117. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, T.; Miyauchi, K. Discovery and characterization of tRNAIle lysidine synthetase (TilS). FEBS Lett. 2010, 584, 272–277. [Google Scholar] [CrossRef] [PubMed]
- Endo, T.; Uratani, B.; Freese, E. Purine salvage pathways of Bacillus subtilis and effect of guanine on growth of GMP reductase mutants. J. Bacteriol. 1983, 155, 169–179. [Google Scholar] [CrossRef] [PubMed]
- Lin, T.-H.; Hu, Y.-N.; Shaw, G.-C. Two enzymes, TilS and HprT can form a complex to function as a transcriptional activator for the cell division protease gene ftsH in Bacillus subtilis. J. Biochem. 2014, 155, 5–16. [Google Scholar] [CrossRef] [PubMed]
- Ul Haq, I.; Müller, P.; Brantl, S. Intermolecular communication in Bacillus subtilis: RNA-RNA, RNA-protein and small protein-protein interactions. Front Mol. Biosci. 2020, 7, 178. [Google Scholar] [CrossRef]
- Serio, A.W.; Pechter, K.B.; Sonenshein, A.L. Bacillus subtilis aconitase is required for efficient late sporulation gene expression. J. Bacteriol. 2006, 188, 6396–6405. [Google Scholar] [CrossRef]
- Pechter, K.B.; Meyer, F.M.; Serio, A.W.; Stülke, J.; Sonenshein, A.L. Two roles for aconitase in the regulation of tricarboxylic acid branch gene expression in Bacillus subtilis. J. Bacteriol. 2013, 195, 1525–1537. [Google Scholar] [CrossRef]
- Bachem, S.; Stülke, J. Regulation of the Bacillus subtilis GlcT antiterminator protein by components of the phosphotransferase system. J. Bacteriol. 1998, 180, 5319–5326. [Google Scholar] [CrossRef]
- Schilling, O.; Langbein, I.; Müller, M.; Schmalisch, M.; Stülke, J. A protein-dependent riboswitch controlling ptsGHI operon expression in Bacillus subtilis: RNA structure rather than sequence provides interaction specificity. Nucleic Acids Res. 2004, 32, 2853–2864. [Google Scholar] [CrossRef]
- Fedorova, K.; Kayumov, A.; Woyda, K.; Ilinskaja, O.; Forchhammer, K. Transcription factor TnrA inhibits the biosynthetic activity of glutamine synthetase in Bacillus subtilis. FEBS Lett. 2013, 587, 1293–1298. [Google Scholar] [CrossRef]
- Wray, L.V., Jr.; Zalieckas, J.M.; Fisher, S.H. Bacillus subtilis glutamine synthetase controls gene expression through a protein-protein interaction with transcription factor TnrA. Cell 2001, 107, 427–435. [Google Scholar] [CrossRef]
- Carpousis, A.J. The RNA degradosome of Escherichia coli: An mRNA-degrading machine assembled on RNase E. Annu. Rev. Microbiol. 2007, 61, 71–87. [Google Scholar] [CrossRef] [PubMed]
- Durand, S.; Tomasini, A.; Braun, F.; Condon, C.; Romby, P. sRNA and mRNA turnover in Gram-positive bacteria. FEMS Microbiol. Rev. 2015, 39, 316–330. [Google Scholar] [CrossRef] [PubMed]
- Shahbabian, K.; Jamalli, A.; Zig, L.; Putzer, H. RNase Y, a novel endoribonuclease, initiates riboswitch turnover in Bacillus subtilis. EMBO J. 2009, 28, 3523–3533. [Google Scholar] [CrossRef] [PubMed]
- Oussenko, I.A.; Abe, T.; Ujiie, H.; Muto, A.; Bechhofer, D.H. Participation of 3′- to -5′ exoribonucleases in the turnover of Bacillus subtilis mRNA. J. Bacteriol. 2005, 187, 2758–2767. [Google Scholar] [CrossRef] [PubMed]
- Even, S.; Pellegrini, O.; Zig, L.; Labas, V.; Vinh, J.; Bréchemmier-Baey, D.; Putzer, H. Ribonucleases J1 and J2: Two novel endoribonucleases in B. subtilis with functional homology to E. coli RNase E. Nucleic Acids Res. 2005, 33, 2145–2152. [Google Scholar] [CrossRef]
- Lehnik-Habrink, M.; Pförtner, H.; Rempeters, L.; Pietack, N.; Herzberg, C.; Stülke, J. The RNA degradosome in Bacillus subtilis: Identification of CshA as the major RNA helicase in the multiprotein complex. Mol. Microbiol. 2010, 77, 958–971. [Google Scholar] [CrossRef]
- Cascante-Estepa, N.; Gunka, K.; Stülke, J. Localization of components of the RNA-degrading machine in Bacillus subtilis. Front. Microbiol. 2016, 7, 1492. [Google Scholar] [CrossRef]
- Newman, J.A.; Hewitt, L.; Rodrigues, C.; Solovyova, A.S.; Harwood, C.R.; Lewis, R.J. Dissection of the network of interactions that links RNA processing with glycolysis in the Bacillus subtilis degradosome. J. Mol Biol. 2012, 416, 121–136. [Google Scholar] [CrossRef]
- Mathy, N.; Hébert, A.; Mervelet, P.; Bénard, L.; Dorléans, A.; Li de la Sierra-Gallay, I.; Noirot, P.; Putzer, H.; Condon, C. Bacillus subtilis ribonucleases J1 and J2 form a complex with altered enzyme behavior. Mol. Microbiol. 2010, 75, 489–498. [Google Scholar] [CrossRef]
- Figaro, S.; Durand, S.; Gilet, L.; Cayet, N.; Sachse, M.; Condon, C. Bacillus subtilis mutants with knockouts of the genes encoding ribonucleases RNase Y and RNase J1 are viable, with major defects in cell morphology, sporulation, and competence. J. Bacteriol. 2013, 195, 2340–2348. [Google Scholar] [CrossRef] [PubMed]
- Dorléans, A.; Li de la Sierra-Gallay, I.; Piton, J.; Zig, L.; Gilet, L.; Putzer, H.; Condon, C. Molecular basis for the recognition and cleavage of RNA by the bifunctional 5′-3′ exo/endoribonuclease RNase J. Structure 2011, 19, 1252–1261. [Google Scholar] [CrossRef]
- Hunt, A.; Rawlins, J.P.; Thomaides, H.B.; Errington, J. Functional analysis of 11 putative essential genes. Microbiology 2006, 152, 2895–2907. [Google Scholar] [CrossRef]
- Mäder, U.; Zig, L.; Kretschmer, J.; Homuth, G.; Putzer, H. mRNA processing by RNases J1 and J2 affects Bacillus subtilis gene expression on a global scale. Mol. Microbiol. 2008, 70, 183–196. [Google Scholar] [CrossRef] [PubMed]
- DeLoughery, A.; Lalanne, J.B.; Losick, R.; Li, G.W. Maturation of polycistronic mRNAs by the endoribonuclease RNase Y and its associated Y-complex in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 2018, 115, E5585–E5594. [Google Scholar] [CrossRef] [PubMed]
- Hamouche, L.; Billaudeau, C.; Rocca, A.; Chastanet, A.; Ngo, S.; Laalami, S.; Putzer, H. Dynamic membrane localization of RNase Y in Bacillus subtilis. mBio 2020, 11, e03335-19. [Google Scholar] [CrossRef]
- Lehnik-Habrink, M.; Schaffer, M.; Mäder, U.; Diethmaier, C.; Herzberg, C.; Stülke, J. RNA processing in Bacillus subtilis: Identification of targets of the essential RNase Y. Mol. Microbiol. 2011, 81, 1459–1473. [Google Scholar] [CrossRef]
- Fillinger, S.; Boschi-Muller, S.; Azza, S.; Dervyn, E.; Branlant, G.; Aymerich, S. Two glyceraldehyde-3-phopshate dehydrogenases with opposite physiological roles in a nonphotosynthetic bacterium. J. Biol. Chem. 2000, 275, 14031–14037. [Google Scholar] [CrossRef]
- Skarzyński, T.; Moody, P.C.; Wonacott, A.J. Structure of holo-glyceraldehyde-3-phosphate dehydrogenase from Bacillus stearothermophilus at 1.8 A resolution. J. Mol. Biol. 1987, 193, 171–187. [Google Scholar] [PubMed]
- Maaß, S.; Wachlin, G.; Bernhardt, J.; Eymann, C.; Fromion, V.; Riedel, K.; Becher, D.; Hecker, M. Highly precise quantification of protein molecules per cell during stress and starvation responses in Bacillus subtilis. Proteomics 2014, 13, 2260–2276. [Google Scholar]
- Gerth, U.; Krieger, E.; Zühlke, D.; Reder, A.; Völker, U.; Hecker, M. Stability of proteins out of service: The GapB case of Bacillus subtilis. J. Bacteriol. 2017, 199, e00148-17. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Meile, J.C.; Wu, L.J.; Ehrlich, S.D.; Errington, J.; Noirot, P. Systematic localisation of proteins fused to the green fluorescent protein in Bacillus subtilis: Identification of new proteins at the DNA replication factory. Proteomics 2006, 6, 2135–2146. [Google Scholar] [CrossRef]
- Hahne, H.; Wolff, S.; Hecker, M.; Becher, D. From complementarity to comprehensiveness—Targeting the membrane proteome of growing Bacillus subtilis by divergent approaches. Proteomics 2008, 8, 4123–4136. [Google Scholar] [CrossRef]
- Matta, S.K.; Agarwal, S.; Bhatnagar, R. Surface localized and extracellular glaceraldehyde-3-phosphate dehydrogenase of Bacillus anthracis is a plasminogen binding protein. Biochim. Biophys. Acta 2010, 1804, 2111–2120. [Google Scholar] [CrossRef] [PubMed]
- Argawal, V.; Hammerschmidt, S.; Malm, S.; Bergmann, S.; Riesbeck, K.; Blom, A.M. Enolase of Streptococcus pneumoniae binds human complement inhibitor C4b-binding protein and contributes to complement evasion. J. Immunol. 2012, 189, 3575–3584. [Google Scholar]
- Jannière, L.; Canceill, D.; Suski, C.; Kanga, S.; Dalmais, B.; Lestini, R.; Monnier, A.F.; Chapuis, J.; Bolotin, A.; Ehrlich, S.D. Genetic evidence for a link between glycolysis and DNA replication. PLoS ONE 2007, 2, e447. [Google Scholar] [CrossRef] [PubMed]
- Evguenieva-Hackenberg, E.; Schiltz, E.; Klug, G. Dehydrogenases from all three domains of life cleave RNA. J. Biol. Chem. 2002, 277, 46145–46150. [Google Scholar] [CrossRef] [PubMed]
- Muntel, J.; Fromion, V.; Goelzer, A.; Maaß, S.; Mäder, U.; Büttner, K.; Hecker, M.; Becher, D. Comprehensive absolute quantification of the cytosolic proteome of Bacillus subtilis based on data independent, parallel fragmentation in liquid chromatography/mass spectrometry (LC/MS(E)). Mol. Cell. Proteom. 2014, 13, 1008–1019. [Google Scholar] [CrossRef]
- Müller, P.; Jahn, N.; Ring, C.; Maiwald, C.; Neubert, R.; Meißner, C.; Brantl, S. A multistress responsive type I toxin-antitoxin system: bsrE/SR5 from the B. subtilis chromosome. RNA Biol. 2016, 13, 511–523. [Google Scholar] [CrossRef]
- Heidrich, N.; Chinali, A.; Gerth, U.; Brantl, S. The small untranslated RNA SR1 from the Bacillus subtilis genome is involved in the regulation of arginine catabolism. Mol. Microbiol. 2006, 62, 520–536. [Google Scholar] [CrossRef]
- Heidrich, N.; Moll, I.; Brantl, S. In vitro analysis of the interaction between the small RNA SR1 and its primary target ahrC mRNA. Nucleic Acids Res. 2007, 35, 4331–4346. [Google Scholar] [CrossRef]
- Daou-Chabo, R.; Condon, C. RNase J1 endonuclease activity as a probe of RNA secondary structure. RNA 2009, 15, 1417–1425. [Google Scholar] [CrossRef] [PubMed]
- Gimpel, M.; Preis, H.; Barth, E.; Gramzow, L.; Brantl, S. SR1—A small RNA with two remarkably conserved functions. Nucleic Acids Res. 2012, 40, 11659–11672. [Google Scholar] [CrossRef] [PubMed]
- Gimpel, M.; Maiwald, C.; Wiedemann, C.; Görlach, M.; Brantl, S. Characterization of the interaction between the small RNA-encoded peptide SR1P and GapA from Bacillus subtilis. Microbiology 2017, 163, 1248–1259. [Google Scholar] [CrossRef] [PubMed]
- Šikova, M.; Wiedermannová, J.; Převorovský, M.; Barvík, I.; Sudzinová, P.; Kofroňová, O.; Benada, O.; Šanderová, H.; Condon, C.; Krásny, L. The torpedo effect in Bacillus subtilis: RNase J1 resolves stalled transcription complexes. EMBO J. 2020, 39, e102500. [Google Scholar] [CrossRef] [PubMed]
- Brewer, J.M.; Ellis, P.D. 31P-NMR studies of the effect of various metals on substrate binding to yeast enolase. J. Inorg. Biochem. 1983, 18, 71–82. [Google Scholar] [CrossRef]
- Leyva-Vazquez, M.A.; Setlow, P. Cloning and nucleotide sequences of the genes encoding triose phosphate isomerase, phosphoglycerate mutase, and enolase from Bacillus subtilis. J. Bacteriol. 1994, 176, 3903–3910. [Google Scholar] [CrossRef]
- Brown, C.K.; Kuhlman, P.L.; Mattingly, S.; Slates, K.; Calie, P.J.; Farrar, W.W. A model of the quarternary structure of enolases, based on structural and evolutionary analysis of the actameric enolase from Bacillus subtilis. J. Protein Chem. 1998, 17, 855–866. [Google Scholar] [CrossRef]
- Jers, C.; Pedersen, M.M.; Paspaliari, D.K.; Schütz, W.; Johnsson, C.; Soufi, B.; Macek, B.; Jensen, P.R.; Mijakovic, I. Bacillus subtilis BY-kinase PtkA controls enzyme activity and localization of its protein substrates. Mol. Microbiol. 2010, 77, 287–299. [Google Scholar] [CrossRef]
- Yang, C.-K.; Ewis, E.H.; Zhang, X.Z.; Lu, C.-D.; Hu, H.-J.; Pan, Y.; Abdelal, A.T.; Tai, P.C. Nonclassical protein secretion by Bacillus subtilis in the stationary phase is not due to cell lysis. J. Bacteriol. 2011, 193, 5607–5615. [Google Scholar] [CrossRef]
- Yang, C.-K.; Zhang, X.Z.; Lu, C.-D.; Tai, P.C. An internal hydrophobic helical domain of Bacillus subtilis enolase is essential but not sufficient as a non-cleavable signal for its secretion. Biochem. Biophys. Res. Commun. 2014, 446, 901–905. [Google Scholar] [CrossRef] [PubMed]
- Kainulainen, V.; Korhonen, T.K. Dancing to another tune—Adhesive moonlighting proteins in bacteria. Biology 2014, 3, 178–204. [Google Scholar] [CrossRef] [PubMed]
- Bergmann, S.; Rohde, M.; Chhatwal, G.S.; Hammerschmidt, S. α-Enolase of Streptococcus pneumoniae is a plasmin(ogen)-binding protein displayed on the bacterial cell surface. Mol. Microbiol. 2001, 40, 1273–1287. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, S.; Kulshreshtha, P.; Mukku, D.B.; Bhatnagar, R. α-Enolase binds to human plasminogen on the surface of Bacillus anthracis. Biochim. Biophys. Acta. 2008, 1784, 986–994. [Google Scholar] [CrossRef] [PubMed]
- Mu, Y.; Xin, Y.; Guo, T.; Kong, J. Identification and characterization of a moonlighting protein-enolase for surface display in Streptococcus thermophilus. Microb. Cell Fact. 2020, 19, 132. [Google Scholar] [CrossRef]
- Mars, R.A.; Mendonça, K.; Denham, E.L.; van Dijl, J.M. The reduction in small ribosomal subunit abundance in ethanol-stressed cells of Bacillus subtilis is mediated by a SigB-dependent antisense RNA. Biochim. Biophys. Acta 2015, 1853, 2553–2559. [Google Scholar] [CrossRef]
- Koch, G.; Wermser, C.; Acosta, I.C.; Kricks, L.; Stengel, S.T.; Yepes, A.; Lopez, D. Attenuating Staphylococcus aureus virulence by targeting flotillin protein scaffold activity. Cell. Chem. Biol. 2017, 24, 845–857. [Google Scholar] [CrossRef]
- Aït-Bara, A.; Carpousis, A.J. RNA degradosomes in bacteria and chloroplasts: Classification, distribution and evolution of RNase E homologs. Mol. Microbiol. 2015, 97, 1021–1135. [Google Scholar] [CrossRef]
- Roux, C.M.; DeMuth, J.P.; Dunman, P.M. Characterization of components of the Staphylococcus aureus mRNA degradosome holoenzyme-like complex. J. Bacteriol. 2011, 193, 5520–5526. [Google Scholar] [CrossRef]
- Redder, P. Molecular and genetic interactions of the RNA degradation machineries in Firmicute bacteria. Wiley Interdiscip. Rev. RNA 2018. [Google Scholar] [CrossRef]
- Khemici, V.; Prados, J.; Linder, P.; Redder, P. Decay-initiating endoribonucleolytic cleavage by RNase Y is kept under tight control via sequence preference and sub-cellular localisation. PLoS Genet. 2015, 11, e1005577. [Google Scholar] [CrossRef] [PubMed]
- Bugrysheva, J.V.; Scott, J.R. The ribonucleases J1 and J2 are essential for growth and have independent roles in mRNA decay in Streptococcus pyogenes. Mol. Microbiol. 2010, 75, 731–743. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ul Haq, I.; Brantl, S. Moonlighting in Bacillus subtilis: The Small Proteins SR1P and SR7P Regulate the Moonlighting Activity of Glyceraldehyde 3-Phosphate Dehydrogenase A (GapA) and Enolase in RNA Degradation. Microorganisms 2021, 9, 1046. https://doi.org/10.3390/microorganisms9051046
Ul Haq I, Brantl S. Moonlighting in Bacillus subtilis: The Small Proteins SR1P and SR7P Regulate the Moonlighting Activity of Glyceraldehyde 3-Phosphate Dehydrogenase A (GapA) and Enolase in RNA Degradation. Microorganisms. 2021; 9(5):1046. https://doi.org/10.3390/microorganisms9051046
Chicago/Turabian StyleUl Haq, Inam, and Sabine Brantl. 2021. "Moonlighting in Bacillus subtilis: The Small Proteins SR1P and SR7P Regulate the Moonlighting Activity of Glyceraldehyde 3-Phosphate Dehydrogenase A (GapA) and Enolase in RNA Degradation" Microorganisms 9, no. 5: 1046. https://doi.org/10.3390/microorganisms9051046
APA StyleUl Haq, I., & Brantl, S. (2021). Moonlighting in Bacillus subtilis: The Small Proteins SR1P and SR7P Regulate the Moonlighting Activity of Glyceraldehyde 3-Phosphate Dehydrogenase A (GapA) and Enolase in RNA Degradation. Microorganisms, 9(5), 1046. https://doi.org/10.3390/microorganisms9051046




