Ecophysiological Features Shape the Distribution of Prophages and CRISPR in Sulfate Reducing Prokaryotes
Abstract
1. Introduction
2. Material and Methods
2.1. Searching for Prophages and CRISPR Elements in the Genomes of SRP
2.2. Ecophysiological Analysis of SRP
2.3. Statistical Analysis
2.4. Experimental Analysis
3. Results and Discussion
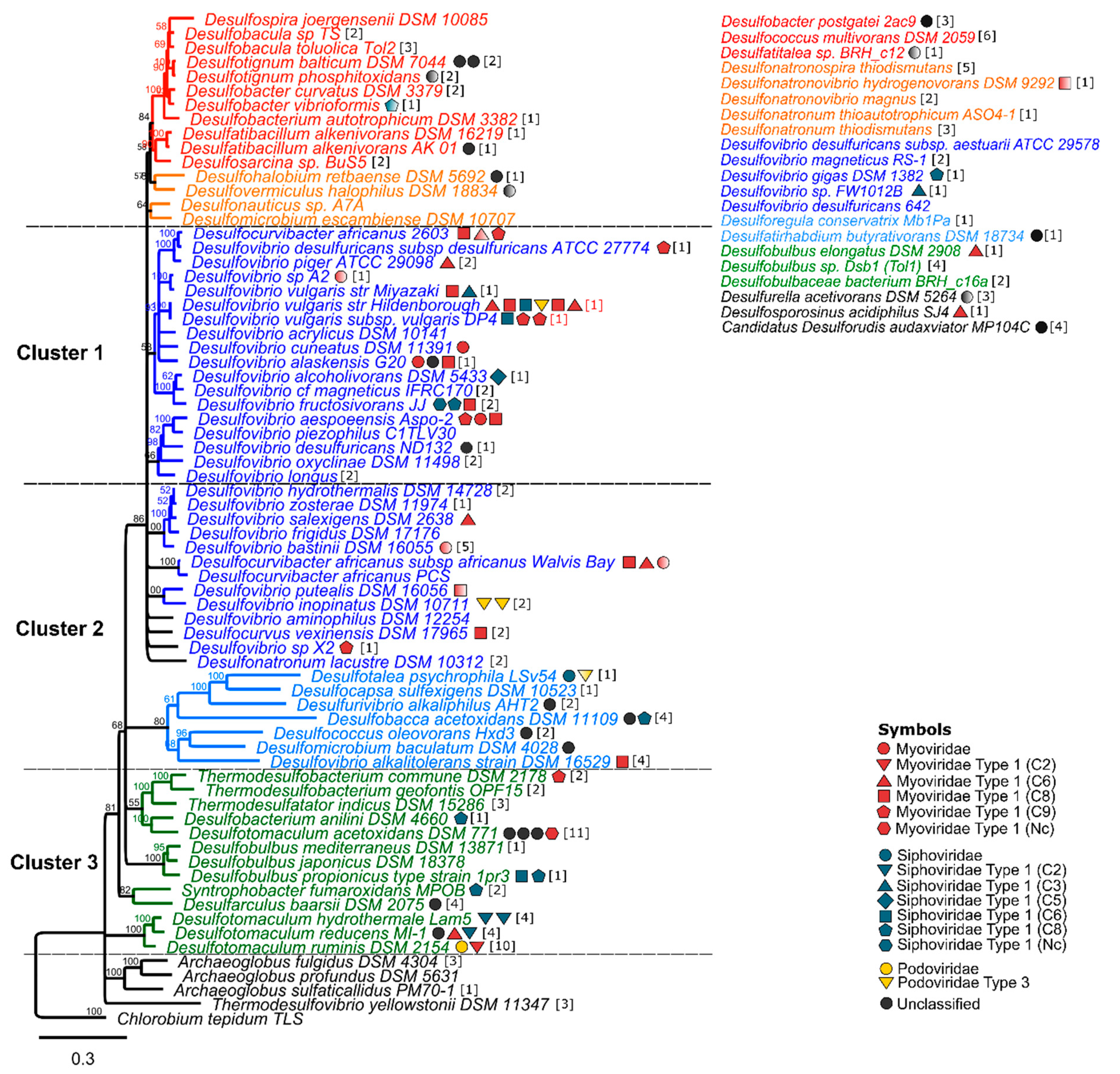
3.1. CRISPR–Cas Systems Are Present in a High Proportion of SRP
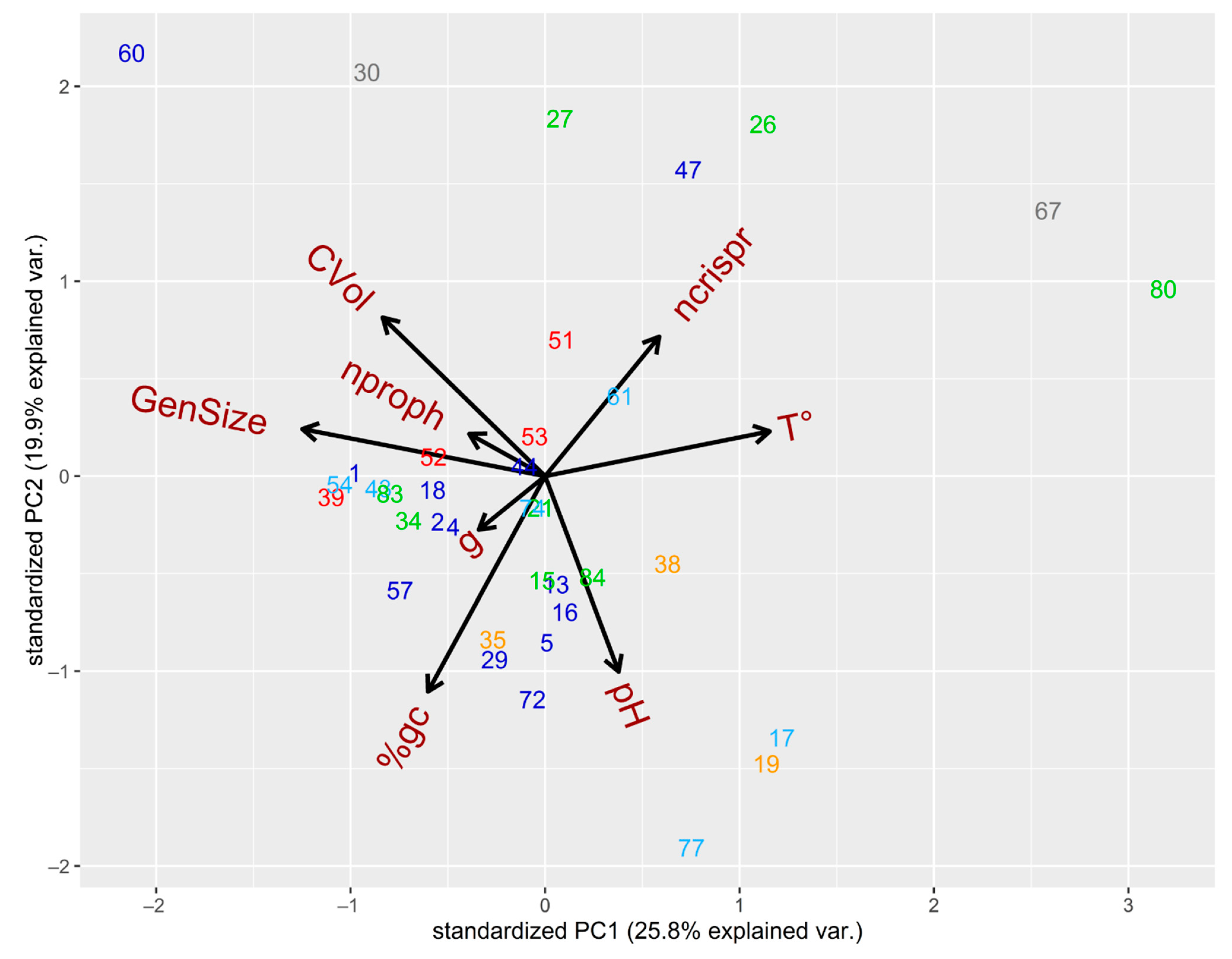
3.2. Ecophysiological Traits Shape the Distribution of Prophages and CRISPR in SRP
3.3. SRP Prophages Are Mainly Myoviridae
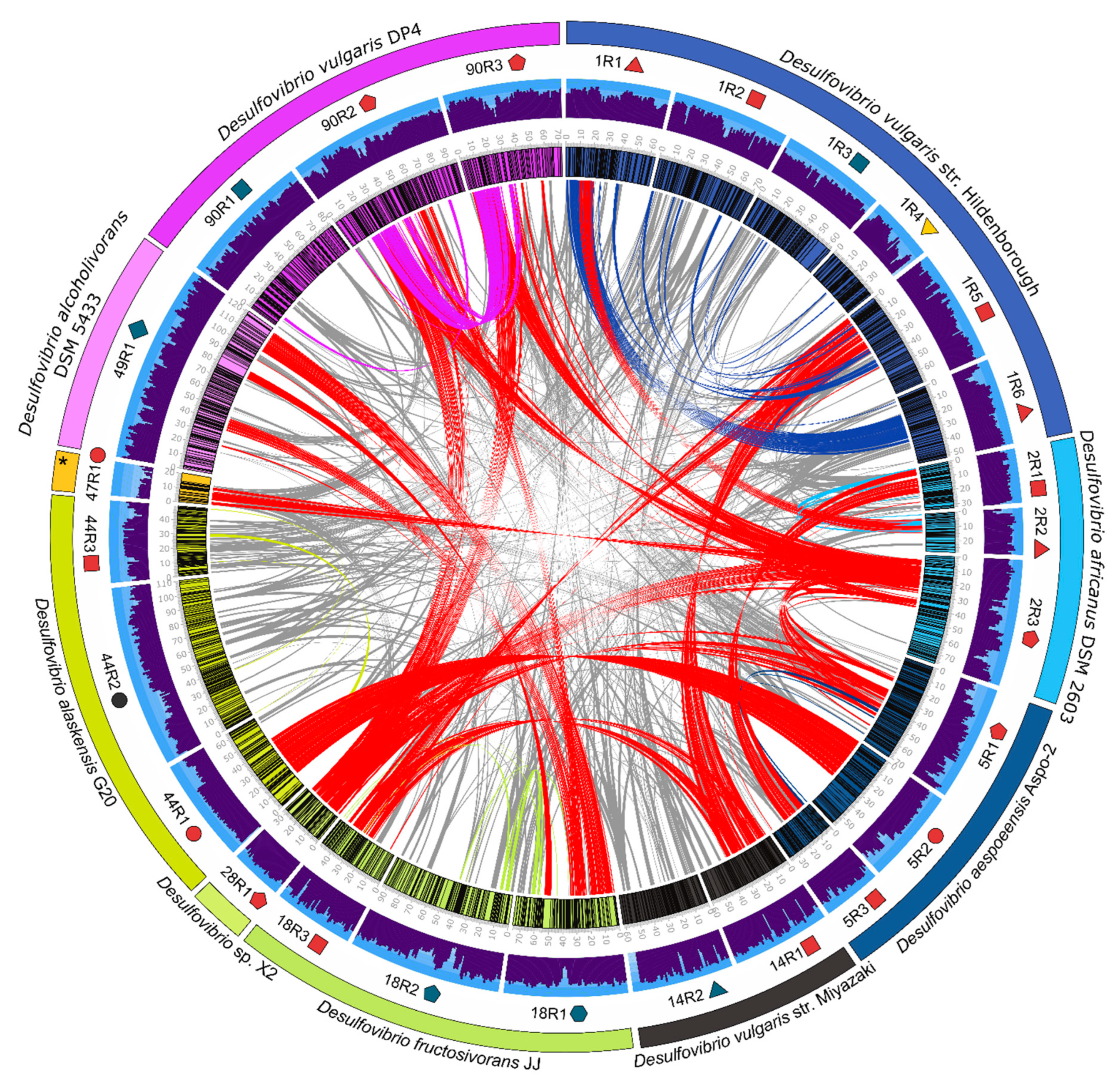
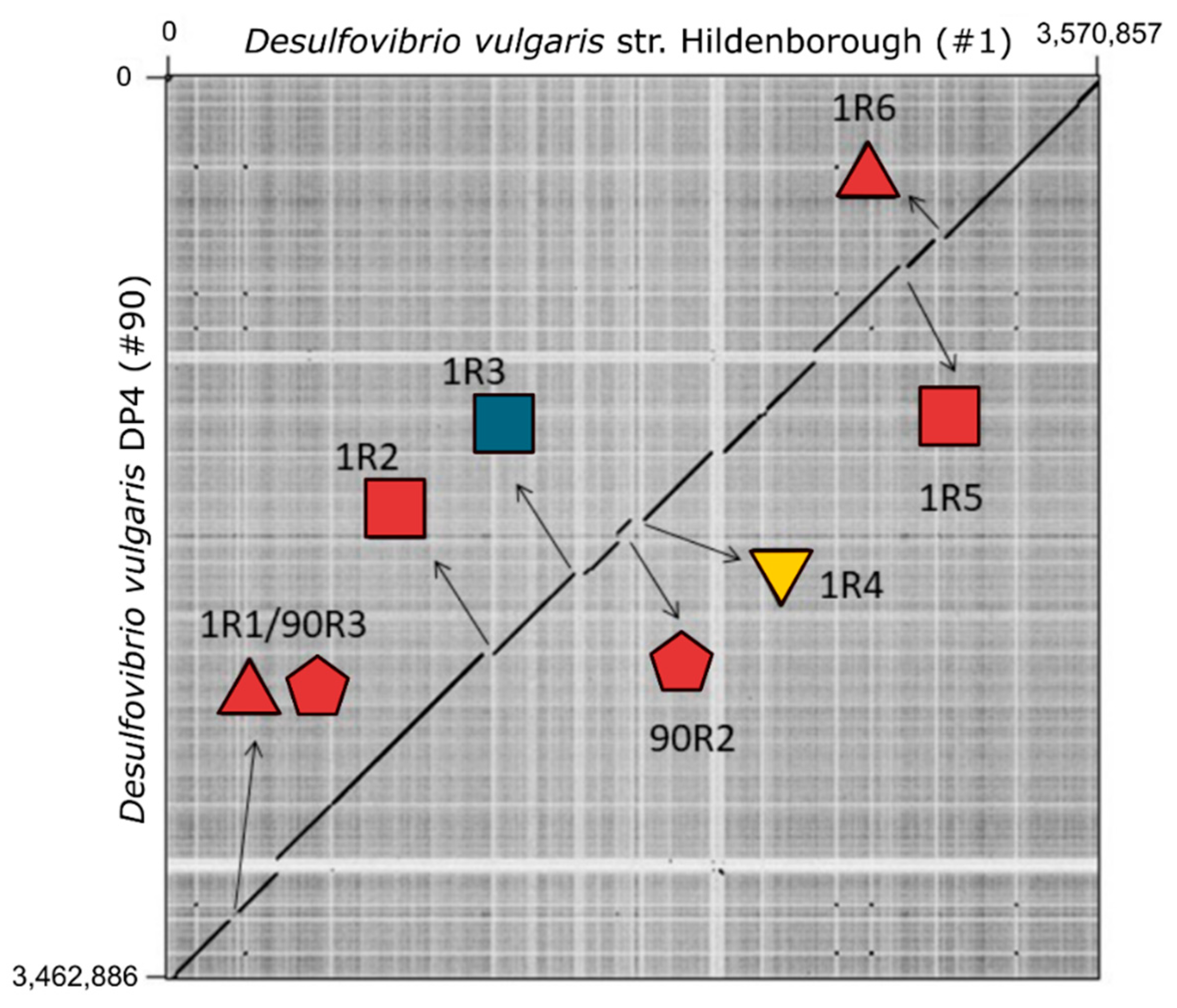
3.4. Desulfovibrio Prophages Exhibited High Degree of Homology
3.5. Induction of Desulfovibrio Prophages
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pedulla, M.L.; Ford, M.E.; Houtz, J.M.; Karthikeyan, T.; Wadsworth, C.; Lewis, J.A.; Jacobs-Sera, D.; Falbo, J.; Gross, J.; Pannunzio, N.R.; et al. Origins of Highly Mosaic Mycobacteriophage Genomes. Cell 2003, 113, 171–182. [Google Scholar] [CrossRef]
- Hatfull, G.F. Dark Matter of the Biosphere: The Amazing World of Bacteriophage Diversity. J. Virol. 2015, 89, 8107–8110. [Google Scholar] [CrossRef] [PubMed]
- Bobay, L.M.; Rocha, E.P.; Touchon, M. The adaptation of temperate bacteriophages to their host genomes. Mol. Biol. Evol. 2013, 30, 737–751. [Google Scholar] [CrossRef]
- Hobbs, Z.; Abedon, S.T. Diversity of phage infection types and associated terminology: The problem with ‘Lytic or lysogenic’. FEMS Microbiol. Lett. 2016, 363. [Google Scholar] [CrossRef] [PubMed]
- Weitz, J.S. Quantitative Viral Ecology. In CHAPTER TWO. Viral Life History Traits; Princeton University Press: Princeton, NJ, USA, 2016; p. 24. [Google Scholar]
- Bobay, L.-M.; Touchon, M.; Rocha, E.P.C. Pervasive domestication of defective prophages by bacteria. Proc. Natl. Acad. Sci. USA 2014, 111, 12127–12132. [Google Scholar] [CrossRef] [PubMed]
- Canchaya, C.; Proux, C.; Fournous, G.; Bruttin, A.; Brüssow, H. Prophage Genomics. Microbiol. Mol. Biol. Rev. 2003, 67, 238–276. [Google Scholar] [CrossRef]
- Sausset, R.; Petit, M.A.; Gaboriau-Routhiau, V.; De Paepe, M. New insights into intestinal phages. Mucosal. Immunol. 2020, 13, 205–215. [Google Scholar] [CrossRef]
- Wang, X.; Kim, Y.; Ma, Q.; Hong, S.H.; Pokusaeva, K.; Sturino, J.M.; Wood, T.K. Cryptic prophages help bacteria cope with adverse environments. Nat. Commun. 2010, 1, 147. [Google Scholar] [CrossRef]
- Clokie, M.R.J.; Millard, A.D.; Letarov, A.V.; Heaphy, S. Phages in nature. Bacteriophage 2011, 1, 31–45. [Google Scholar] [CrossRef]
- Thompson, L.R.; Zeng, Q.; Kelly, L.; Huang, K.H.; Singer, A.U.; Stubbe, J.; Chisholm, S.W. Phage auxiliary metabolic genes and the redirection of cyanobacterial host carbon metabolism. Proc. Natl. Acad. Sci. USA 2011, 108, E757. [Google Scholar] [CrossRef]
- Crummett, L.T.; Puxty, R.J.; Weihe, C.; Marston, M.F.; Martiny, J.B.H. The genomic content and context of auxiliary metabolic genes in marine cyanomyoviruses. Virology 2016, 499, 219–229. [Google Scholar] [CrossRef]
- Gibson, G.R. Physiology and ecology of the sulphate-reducing bacteria. J. Appl. Bacteriol. 1990, 69, 769–797. [Google Scholar] [CrossRef] [PubMed]
- Postgate, J.R. Recent advances in the study of the sulfate-reducing bacteria. Bacteriol. Rev. 1965, 29, 425–441. [Google Scholar] [CrossRef] [PubMed]
- Mussmann, M.; Richter, M.; Lombardot, T.; Meyerdierks, A.; Kuever, J.; Kube, M.; Glöckner, F.O.; Amann, R. Clustered Genes Related to Sulfate Respiration in Uncultured Prokaryotes Support the Theory of Their Concomitant Horizontal Transfer. J. Bacteriol. 2005, 187, 7126–7137. [Google Scholar] [CrossRef]
- Muyzer, G.; Stams, A.J. The ecology and biotechnology of sulphate-reducing bacteria. Nat. Rev. Microbiol 2008, 6, 441–454. [Google Scholar] [CrossRef]
- Jørgensen, B.B. The sulfur cycle of a coastal marine sediment (Limfjorden, Denmark). Limnol. Oceanogr. 1977, 22, 814–832. [Google Scholar] [CrossRef]
- Bak, F.; Pfennig, N. Microbial sulfate reduction in littoral sediment of Lake Constance. FEMS Microbiol. Lett. 1991, 85, 31–42. [Google Scholar] [CrossRef]
- Castro, H.; Reddy, K.R.; Ogram, A. Composition and Function of Sulfate-Reducing Prokaryotes in Eutrophic and Pristine Areas of the Florida Everglades. Appl. Environ. Microbiol. 2002, 68, 6129–6137. [Google Scholar] [CrossRef] [PubMed]
- Handley, J.; Adams, V.; Akagi, J.M. Morphology of bacteriophage-like particles from Desulfovibrio vulgaris. J. Bacteriol. 1973, 115, 1205–1207. [Google Scholar] [CrossRef]
- Seyedirashti, S.; Wood, C.; Akagi, J.M. Induction and partial purification of bacteriophages from Desulfovibrio vulgaris (Hildenborough) and Desulfovibrio desulfuricans ATCC 13541. J. Gen. Microbiol. 1991, 137, 1545–1549. [Google Scholar] [CrossRef]
- Seyedirashti, S.; Wood, C.; Akagi, J.M. Molecular characterization of two bacteriophages isolated from Desulfovibrio vulgaris NCIMB 8303 (Hildenborough). J. Gen. Microbiol. 1992, 138, 1393–1397. [Google Scholar] [CrossRef]
- Walker, C.B.; Stolyar, S.; Chivian, D.; Pinel, N.; Gabster, J.A.; Dehal, P.S.; He, Z.; Yang, Z.K.; Yen, H.-C.B.; Zhou, J.; et al. Contribution of mobile genetic elements to Desulfovibrio vulgaris genome plasticity. Environ. Microbiol. 2009, 11, 2244–2252. [Google Scholar] [CrossRef] [PubMed]
- Walker, C.B.; Stolyar, S.S.; Pinel, N.; Yen, H.C.; He, Z.; Zhou, J.; Wall, J.D.; Stahl, D.A. Recovery of temperate Desulfovibrio vulgaris bacteriophage using a novel host strain. Environ. Microbiol 2006, 8, 1950–1959. [Google Scholar] [CrossRef] [PubMed]
- Kamimura, K.; Araki, M. Isolation and Characterization of a Bacteriophage Lytic for Desulfovibrio salexigens, a Salt-Requiring, Sulfate-Reducing Bacterium. Appl. Environ. Microbiol. 1989, 55, 645–648. [Google Scholar] [CrossRef] [PubMed]
- Eydal, H.S.; Jagevall, S.; Hermansson, M.; Pedersen, K. Bacteriophage lytic to Desulfovibrio aespoeensis isolated from deep groundwater. ISME J. 2009, 3, 1139–1147. [Google Scholar] [CrossRef] [PubMed]
- Costeira, R.; Doherty, R.; Allen, C.C.R.; Larkin, M.J.; Kulakov, L.A. Analysis of viral and bacterial communities in groundwater associated with contaminated land. Sci. Total Environ. 2019, 656, 1413–1426. [Google Scholar] [CrossRef]
- Fouts, D.E. Phage_Finder: Automated identification and classification of prophage regions in complete bacterial genome sequences. Nucleic Acids Res. 2006, 34, 5839–5851. [Google Scholar] [CrossRef] [PubMed]
- Roux, S.; Enault, F.; Hurwitz, B.L.; Sullivan, M.B. VirSorter: Mining viral signal from microbial genomic data. PeerJ 2015, 3, e985. [Google Scholar] [CrossRef]
- Arndt, D.; Grant, J.R.; Marcu, A.; Sajed, T.; Pon, A.; Liang, Y.; Wishart, D.S. PHASTER: A better, faster version of the PHAST phage search tool. Nucleic Acids Res. 2016, 44, W16–W21. [Google Scholar] [CrossRef] [PubMed]
- Crispim, J.S.; Dias, R.S.; Vidigal, P.M.P.; de Sousa, M.P.; da Silva, C.C.; Santana, M.F.; de Paula, S.O. Screening and characterization of prophages in Desulfovibrio genomes. Sci. Rep. 2018, 8, 9273. [Google Scholar] [CrossRef]
- Akhter, S.; Aziz, R.K.; Edwards, R.A. PhiSpy: A novel algorithm for finding prophages in bacterial genomes that combines similarity- and composition-based strategies. Nucleic Acids Res. 2012, 40, e126. [Google Scholar] [CrossRef]
- Ravin, V.; Ravin, N.; Casjens, S.; Ford, M.E.; Hatfull, G.F.; Hendrix, R.W. Genomic sequence and analysis of the atypical temperate bacteriophage N15. J. Mol. Biol. 2000, 299, 53–73. [Google Scholar] [CrossRef]
- Feiner, R.; Argov, T.; Rabinovich, L.; Sigal, N.; Borovok, I.; Herskovits, A.A. A new perspective on lysogeny: Prophages as active regulatory switches of bacteria. Nat. Rev. Microbiol 2015, 13, 641–650. [Google Scholar] [CrossRef]
- Deutsch, D.R.; Utter, B.; Verratti, K.J.; Sichtig, H.; Tallon, L.J.; Fischetti, V.A. Extra-Chromosomal DNA Sequencing Reveals Episomal Prophages Capable of Impacting Virulence Factor Expression in Staphylococcus aureus. Front. Microbiol. 2018, 9. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef] [PubMed]
- Hyatt, D.; Chen, G.-L.; Locascio, P.F.; Land, M.L.; Larimer, F.W.; Hauser, L.J. Prodigal: Prokaryotic gene recognition and translation initiation site identification. BMC Bioinform. 2010, 11, 119. [Google Scholar] [CrossRef] [PubMed]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 2015, 12, 59–60. [Google Scholar] [CrossRef]
- Lopes, A.; Tavares, P.; Petit, M.-A.; Guérois, R.; Zinn-Justin, S. Automated classification of tailed bacteriophages according to their neck organization. BMC Genom. 2014, 15, 1027. [Google Scholar] [CrossRef] [PubMed]
- Hagberg, A.; Swart, P.; Chult, D.S. Exploring Network Structure, Dynamics, and Function Using Networkx; Los Alamos National Lab. (LANL): Los Alamos, NM, USA, 2008. [Google Scholar]
- Krumsiek, J.; Arnold, R.; Rattei, T. Gepard: A rapid and sensitive tool for creating dotplots on genome scale. Bioinformatics 2007, 23, 1026–1028. [Google Scholar] [CrossRef]
- Krzywinski, M.I.; Schein, J.E.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009. [Google Scholar] [CrossRef]
- Couvin, D.; Bernheim, A.; Toffano-Nioche, C.; Touchon, M.; Michalik, J.; Néron, B.; Rocha, E.P.C.; Vergnaud, G.; Gautheret, D.; Pourcel, C. CRISPRCasFinder, an update of CRISRFinder, includes a portable version, enhanced performance and integrates search for Cas proteins. Nucleic Acids Res. 2018, 46, W246–W251. [Google Scholar] [CrossRef] [PubMed]
- Grissa, I.; Vergnaud, G.; Pourcel, C. CRISPRFinder: A web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res. 2007, 35, W52–W57. [Google Scholar] [CrossRef] [PubMed]
- Johnson, R.A.; Wichern, D.W. Applied Multivariate Statistical Analysis; Pearson Prentice Hall: Upper Saddle River, NJ, USA, 2007. [Google Scholar]
- Chang, D.; Keinan, A. Principal Component Analysis Characterizes Shared Pathogenetics from Genome-Wide Association Studies. PLoS Comput. Biol. 2014, 10, e1003820. [Google Scholar] [CrossRef] [PubMed]
- Welch, B.L. The generalisation of student’s problems when several different population variances are involved. Biometrika 1947, 34, 28–35. [Google Scholar] [CrossRef] [PubMed]
- Derrick, B.; Russ, B.; Toher, D.; White, P. Test statistics for the comparison of means for two samples which include both paired observations and independent observations. J. Mod. Appl. Stat. Methods 2017, 16, 137–157. [Google Scholar] [CrossRef]
- Rey, D.; Neuhäuser, M. Wilcoxon-Signed-Rank Test. Int. Encycl. Stat. Sci. 2011, 1658–1659. [Google Scholar] [CrossRef]
- Neuhäuser, M. Wilcoxon–Mann–Whitney Test. In International Encyclopedia of Statistical Science; Springer: Berlin/Heidelberg, Germany, 2011; pp. 1656–1658. [Google Scholar]
- R Development Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2010. [Google Scholar]
- Pfennig, N.; Widdel, F.; Trüper, H.G. The Dissimilatory Sulfate-Reducing Bacteria. In The Prokaryotes: A Handbook on Habitats, Isolation, and Identification of Bacteria; Starr, M.P., Stolp, H., Trüper, H.G., Balows, A., Schlegel, H.G., Eds.; Springer: Berlin/Heidelberg, Germany, 1981; pp. 926–940. [Google Scholar]
- Mukhopadhyay, A.; He, Z.; Alm, E.J.; Arkin, A.P.; Baidoo, E.E.; Borglin, S.C.; Chen, W.; Hazen, T.C.; He, Q.; Holman, H.-Y.; et al. Salt stress in Desulfovibrio vulgaris Hildenborough: An integrated genomics approach. J. Bacteriol. 2006, 188, 4068. [Google Scholar] [CrossRef]
- Touchon, M.; Bernheim, A.; Rocha, E.P. Genetic and life-history traits associated with the distribution of prophages in bacteria. ISME J. 2016, 10, 2744–2754. [Google Scholar] [CrossRef]
- Müller, A.L.; Kjeldsen, K.U.; Rattei, T.; Pester, M.; Loy, A. Phylogenetic and environmental diversity of DsrAB-type dissimilatory (bi)sulfite reductases. ISME J. 2015, 9, 1152–1165. [Google Scholar] [CrossRef] [PubMed]
- Feio, M.J.; Zinkevich, V.; Beech, I.B.; Llobet-Brossa, E.; Eaton, P.; Schmitt, J.; Guezennec, J. Desulfovibrio alaskensis sp. nov., a sulphate-reducing bacterium from a soured oil reservoir. Int. J. Syst. Evol. Microbiol. 2004, 54, 1747–1752. [Google Scholar] [CrossRef]
- Campbell, L.L.; Kasprzycki, M.A.; Postgate, J.R. Desulfovibrio africans sp. n., a new dissimilatory sulfate-reducing bacterium. J. Bacteriol. 1966, 92, 1122–1127. [Google Scholar] [CrossRef]
- Zaarur, S.; Wang, D.T.; Ono, S.; Bosak, T. Influence of Phosphorus and Cell Geometry on the Fractionation of Sulfur Isotopes by Several Species of Desulfovibrio during Microbial Sulfate Reduction. Front. Microbiol. 2017, 8. [Google Scholar] [CrossRef]
- Knoblauch, C.; Sahm, K.; Jørgensen, B.B. Psychrophilic sulfate-reducing bacteria isolated from permanently cold Arctic marine sediments: Description of Desulfofrigus oceanense gen. nov., sp. nov., Desulfofrigus fragile sp. nov., Desulfofaba gelida gen. nov., sp. nov., Desulfotalea psychrophila gen. nov., sp. nov. and Desulfotalea arctica sp. nov. Int. J. Syst. Evol. Microbiol. 1999, 49, 1631–1643. [Google Scholar] [CrossRef]
- Oude Elferink, S.J.; Akkermans-van Vliet, W.M.; Bogte, J.J.; Stams, A.J. Desulfobacca acetoxidans gen. nov., sp. nov., a novel acetate-degrading sulfate reducer isolated from sulfidogenic granular sludge. Int. J. Syst. Bacteriol. 1999, 49 Pt 2, 345–350. [Google Scholar] [CrossRef] [PubMed]
- Zeikus, J.; Dawson, M.; Thompson, T.E.; Ingvorsen, K.; Hatchikian, E.C. Microbial Ecology of Volcanic Sulphidogenesis: Isolation and Characterization of Thermodesulfobacterium commune gen. nov. and sp. nov. Microbiology 1983, 129, 1159–1169. [Google Scholar] [CrossRef]
- Haouari, O.; Fardeau, M.L.; Cayol, J.L.; Casiot, C.; Elbaz-Poulichet, F.; Hamdi, M.; Joseph, M.; Ollivier, B. Desulfotomaculum hydrothermale sp. nov., a thermophilic sulfate-reducing bacterium isolated from a terrestrial Tunisian hot spring. Int. J. Syst. Evol. Microbiol. 2008, 58, 2529–2535. [Google Scholar] [CrossRef] [PubMed]
- Ackermann, H.W. 5500 Phages examined in the electron microscope. Arch. Virol. 2007, 152, 227–243. [Google Scholar] [CrossRef]
- Junier, P.; Vecchia, E.D.; Bernier-Latmani, R. The Response of Desulfotomaculum reducens MI-1 to U(VI) Exposure: A Transcriptomic Study. Geomicrobiol. J. 2011, 28, 483–496. [Google Scholar] [CrossRef][Green Version]
- Widdel, F.; Pfennig, N. Studies on dissimilatory sulfate-reducing bacteria that decompose fatty acids II. Incomplete oxidation of propionate by Desulfobulbus propionicus gen. nov., sp. nov. Arch. Microbiol. 1982, 131, 360–365. [Google Scholar] [CrossRef]
- Pagani, I.; Lapidus, A.; Nolan, M.; Lucas, S.; Hammon, N.; Deshpande, S.; Cheng, J.-F.; Chertkov, O.; Davenport, K.; Tapia, R.; et al. Complete genome sequence of Desulfobulbus propionicus type strain (1pr3). Stand. Genom. Sci. 2011, 4, 100–110. [Google Scholar] [CrossRef]
- McInerney, M.J.; Sieber, J.R.; Gunsalus, R.P. Syntrophy in anaerobic global carbon cycles. Curr. Opin. Biotechnol. 2009, 20, 623–632. [Google Scholar] [CrossRef]
- Hidalgo-Ahumada, C.A.P.; Nobu, M.K.; Narihiro, T.; Tamaki, H.; Liu, W.T.; Kamagata, Y.; Stams, A.J.M.; Imachi, H.; Sousa, D.Z. Novel energy conservation strategies and behaviour of Pelotomaculum schinkii driving syntrophic propionate catabolism. Environ. Microbiol. 2018, 20, 4503–4511. [Google Scholar] [CrossRef]
- Sedano-Núñez, V.T.; Boeren, S.; Stams, A.J.M.; Plugge, C.M. Comparative proteome analysis of propionate degradation by Syntrophobacter fumaroxidans in pure culture and in coculture with methanogens. Environ. Microbiol. 2018, 20, 1842–1856. [Google Scholar] [CrossRef] [PubMed]
- Barrangou, R.; Fremaux, C.; Deveau, H.; Richards, M.; Boyaval, P.; Moineau, S.; Romero, D.A.; Horvath, P. CRISPR Provides Acquired Resistance Against Viruses in Prokaryotes. Science 2007, 315, 1709. [Google Scholar] [CrossRef] [PubMed]
- Bikard, D.; Hatoum-Aslan, A.; Mucida, D.; Marraffini, L.A. CRISPR interference can prevent natural transformation and virulence acquisition during in vivo bacterial infection. Cell Host Microbe 2012, 12, 177–186. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Sanchez, M.J.; Sauvage, E.; Da Cunha, V.; Clermont, D.; Ratsima Hariniaina, E.; Gonzalez-Zorn, B.; Poyart, C.; Rosinski-Chupin, I.; Glaser, P. The highly dynamic CRISPR1 system of Streptococcus agalactiae controls the diversity of its mobilome. Mol. Microbiol. 2012, 85, 1057–1071. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Lv, L.; Wang, X.; Xiu, Z.; Chen, G. Comparative analysis of CRISPR-Cas systems in Klebsiella genomes. J. Basic Microbiol. 2017, 57, 325–336. [Google Scholar] [CrossRef]
- Medina-Aparicio, L.; Dávila, S.; Rebollar-Flores, J.E.; Calva, E.; Hernández-Lucas, I. The CRISPR-Cas system in Enterobacteriaceae. Pathog. Dis. 2018, 76. [Google Scholar] [CrossRef]
- Godde, J.S.; Bickerton, A. The repetitive DNA elements called CRISPRs and their associated genes: Evidence of horizontal transfer among prokaryotes. J. Mol. Evol. 2006, 62, 718–729. [Google Scholar] [CrossRef]
- Watson, B.N.J.; Staals, R.H.J.; Fineran, P.C. CRISPR-Cas-Mediated Phage Resistance Enhances Horizontal Gene Transfer by Transduction. mBio 2018, 9, e02406–e02417. [Google Scholar] [CrossRef]
- Gao, N.L.; Chen, J.; Wang, T.; Lercher, M.J.; Chen, W.-H. Prokaryotic Genome Expansion Is Facilitated by Phages and Plasmids but Impaired by CRISPR. Front. Microbiol. 2019, 10. [Google Scholar] [CrossRef]
- Bradde, S.; Nourmohammad, A.; Goyal, S.; Balasubramanian, V. The size of the immune repertoire of bacteria. Proc. Natl. Acad. Sci. USA 2020, 117, 5144–5151. [Google Scholar] [CrossRef]
- Goldberg, G.W.; McMillan, E.A.; Varble, A.; Modell, J.W.; Samai, P.; Jiang, W.; Marraffini, L.A. Incomplete prophage tolerance by type III-A CRISPR-Cas systems reduces the fitness of lysogenic hosts. Nat. Commun. 2018, 9, 61. [Google Scholar] [CrossRef]
- Hirooka, S.; Hirose, Y.; Kanesaki, Y.; Higuchi, S.; Fujiwara, T.; Onuma, R.; Era, A.; Ohbayashi, R.; Uzuka, A.; Nozaki, H.; et al. Acidophilic green algal genome provides insights into adaptation to an acidic environment. Proc. Natl. Acad. Sci. USA 2017, 114, E8304–E8313. [Google Scholar] [CrossRef] [PubMed]
- Pate, J.L.; Petzold, S.J.; Umbreit, T.H. Two flagellotropic phages and one pilus-specific phage active against Asticcacaulis biprosthecum. Virology 1979, 94, 24–37. [Google Scholar] [CrossRef]
- Gonzalez, F.; Helm, R.F.; Broadway, K.M.; Scharf, B.E. More than Rotating Flagella: Lipopolysaccharide as a Secondary Receptor for Flagellotropic Phage 7-7-1. J. Bacteriol. 2018, 200. [Google Scholar] [CrossRef]
- Phothaworn, P.; Dunne, M.; Supokaivanich, R.; Ong, C.; Lim, J.; Taharnklaew, R.; Vesaratchavest, M.; Khumthong, R.; Pringsulaka, O.; Ajawatanawong, P.; et al. Characterization of Flagellotropic, Chi-Like Salmonella Phages Isolated from Thai Poultry Farms. Viruses 2019, 11, 520. [Google Scholar] [CrossRef] [PubMed]
- Guerrero-Ferreira, R.C.; Viollier, P.H.; Ely, B.; Poindexter, J.S.; Georgieva, M.; Jensen, G.J.; Wright, E.R. Alternative mechanism for bacteriophage adsorption to the motile bacterium Caulobacter crescentus. Proc. Natl. Acad. Sci. USA 2011, 108, 9963–9968. [Google Scholar] [CrossRef] [PubMed]
- Filippini, M.; Middelboe, M. Viral abundance and genome size distribution in the sediment and water column of marine and freshwater ecosystems. FEMS Microbiol. Ecol. 2007, 60, 397–410. [Google Scholar] [CrossRef]
- Danovaro, R.; Corinaldesi, C.; Filippini, M.; Fischer, U.R.; Gessner, M.O.; Jacquet, S.; Magagnini, M.; Velimirov, B. Viriobenthos in freshwater and marine sediments: A review. Freshw. Biol. 2008, 53, 1186–1213. [Google Scholar] [CrossRef]
- Middelboe, M.; Jacquet, S.; Weinbauer, M. Viruses in freshwater ecosystems: An introduction to the exploration of viruses in new aquatic habitats. Freshw. Biol. 2008, 53, 1069–1075. [Google Scholar] [CrossRef]
- Shmakov, S.A.; Sitnik, V.; Makarova, K.S.; Wolf, Y.I.; Severinov, K.V.; Koonin, E.V. The CRISPR Spacer Space Is Dominated by Sequences from Species-Specific Mobilomes. mBio 2017, 8, e01397-17. [Google Scholar] [CrossRef]
- Sandaa, R.-A.; Gómez-Consarnau, L.; Pinhassi, J.; Riemann, L.; Malits, A.; Weinbauer, M.G.; Gasol, J.M.; Thingstad, T.F. Viral control of bacterial biodiversity–evidence from a nutrient-enriched marine mesocosm experiment. Environ. Microbiol. 2009, 11, 2585–2597. [Google Scholar] [CrossRef]
- Sandaa, R.A. Viruses, Environmental. In Encyclopedia of Microbiology, 3rd ed.; Schaechter, M., Ed.; Academic Press: Oxford, UK, 2009; pp. 553–567. [Google Scholar]
- Rapp, B.J.; Wall, J.D. Genetic Transfer in Desulfovibrio desulfuricans. Proc. Natl. Acad. Sci. USA 1987, 84, 9128–9130. [Google Scholar] [CrossRef] [PubMed]
- Crispim, J.S.; Dias, R.S.; Laguardia, C.N.; Araújo, L.C.; da Silva, J.D.; Vidigal, P.M.P.; de Sousa, M.P.; da Silva, C.C.; Santana, M.F.; de Paula, S.O. Desulfovibrio alaskensis prophages and their possible involvement in the horizontal transfer of genes by outer membrane vesicles. Gene 2019, 703, 50–57. [Google Scholar] [CrossRef]
- Mozaheb, N.; Mingeot-Leclercq, M.-P. Membrane Vesicle Production as a Bacterial Defense against Stress. Front. Microbiol. 2020, 11. [Google Scholar] [CrossRef]
- Hernández, S.; Vives, M.J. Phages in Anaerobic Systems. Viruses 2020, 12, 1091. [Google Scholar] [CrossRef]
- Kuever, J.; Rainey, F.A.; Widdel, F. Desulfovibrio. In Bergey’s Manual of Systematics of Archaea and Bacteria; Wiley: New York, NY, USA, 2015; pp. 1–17. [Google Scholar]
- Pace, M.L.; Cole, J.J. Comparative and experimental approaches to top-down and bottom-up regulation of bacteria. Microb. Ecol. 1994, 28, 181–193. [Google Scholar] [CrossRef]
- Holmes, D.E.; Giloteaux, L.; Chaurasia, A.K.; Williams, K.H.; Luef, B.; Wilkins, M.J.; Wrighton, K.C.; Thompson, C.A.; Comolli, L.R.; Lovley, D.R. Evidence of Geobacter-associated phage in a uranium-contaminated aquifer. ISME J. 2015, 9, 333–346. [Google Scholar] [CrossRef]
- Young, K.D. The selective value of bacterial shape. Microbiol. Mol. Biol. Rev. 2006, 70, 660–703. [Google Scholar] [CrossRef] [PubMed]
- Postgate, J. Breathless niches. Nature 1995, 377, 26. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Orellana, R.; Arancibia, A.; Badilla, L.; Acosta, J.; Arancibia, G.; Escar, R.; Ferrada, G.; Seeger, M. Ecophysiological Features Shape the Distribution of Prophages and CRISPR in Sulfate Reducing Prokaryotes. Microorganisms 2021, 9, 931. https://doi.org/10.3390/microorganisms9050931
Orellana R, Arancibia A, Badilla L, Acosta J, Arancibia G, Escar R, Ferrada G, Seeger M. Ecophysiological Features Shape the Distribution of Prophages and CRISPR in Sulfate Reducing Prokaryotes. Microorganisms. 2021; 9(5):931. https://doi.org/10.3390/microorganisms9050931
Chicago/Turabian StyleOrellana, Roberto, Alejandra Arancibia, Leonardo Badilla, Jonathan Acosta, Gabriela Arancibia, Rodrigo Escar, Gustavo Ferrada, and Michael Seeger. 2021. "Ecophysiological Features Shape the Distribution of Prophages and CRISPR in Sulfate Reducing Prokaryotes" Microorganisms 9, no. 5: 931. https://doi.org/10.3390/microorganisms9050931
APA StyleOrellana, R., Arancibia, A., Badilla, L., Acosta, J., Arancibia, G., Escar, R., Ferrada, G., & Seeger, M. (2021). Ecophysiological Features Shape the Distribution of Prophages and CRISPR in Sulfate Reducing Prokaryotes. Microorganisms, 9(5), 931. https://doi.org/10.3390/microorganisms9050931






