The Population Structure of Borrelia lusitaniae Is Reflected by a Population Division of Its Ixodes Vector
Abstract
:1. Introduction
2. Materials and Methods
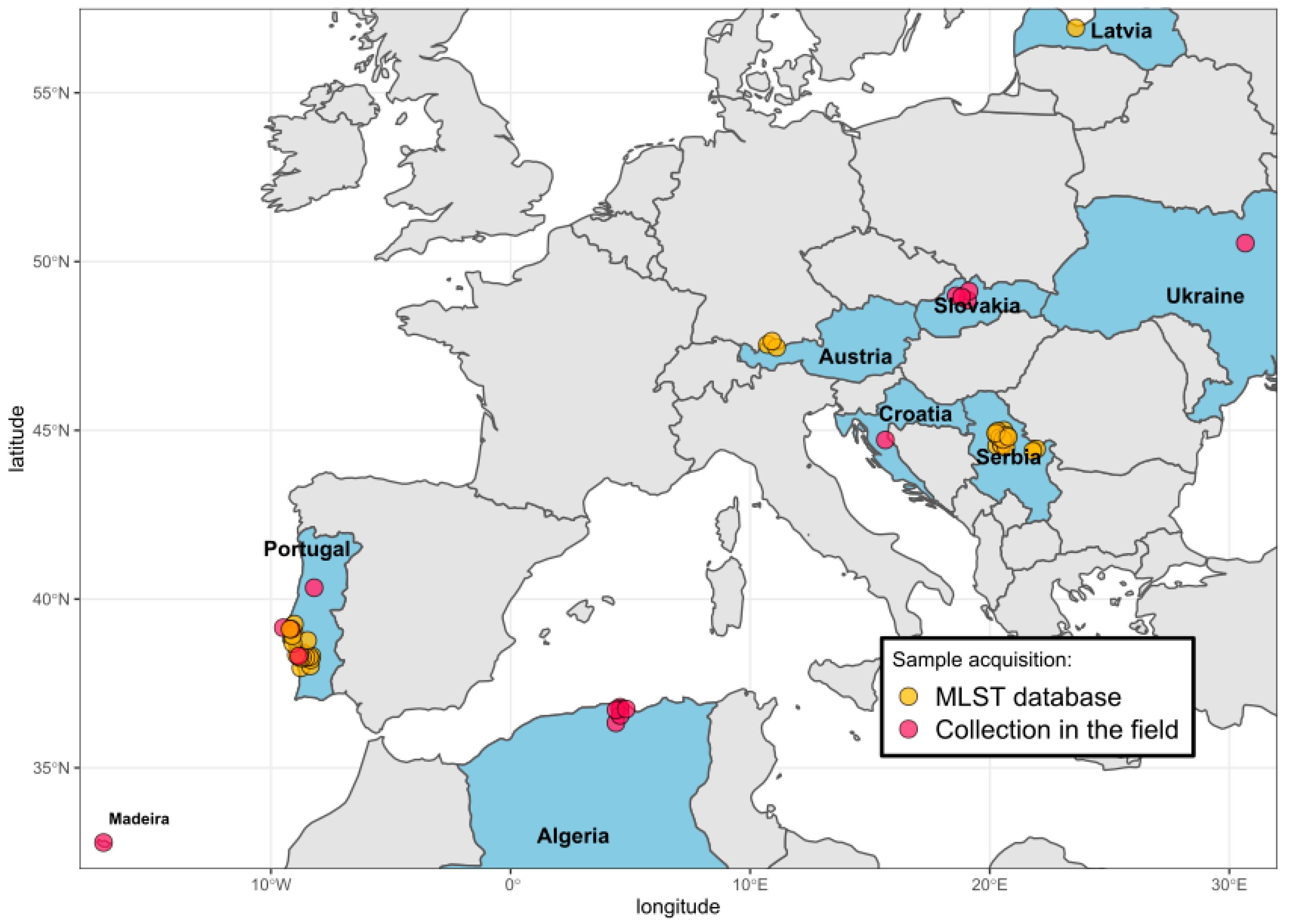
2.1. Tick Collection and Processing
2.2. DNA Extraction
2.3. PCR Borrelia Genes
2.4. PCR on Tick Genes
2.5. Bioinformatic Analysis Recombination and Network Analysis on MLST Genes of Borrelia Samples
2.6. Hierarchical Population Structure Analysis (HPSA) of Borrelia
2.7. Hierarchical Clustering of MLST Genes of Borrelia Samples
2.8. GoeBURST Analysis of Borrelia
2.9. Maximum Likelihood Phylogenies of Tick Samples
3. Results
3.1. Identification of Borrelia lusitaniae in Ticks
3.2. MLST, goeBURST and Phylogenetic Analyses of Borrelia Samples
3.2.1. Multilocus Sequence Typing (MLST)
3.2.2. Clustering Analysis of B. lusitaniae
3.2.3. goeBURST Analysis
3.2.4. Sequence Analysis
3.3. Analysis of Tick Samples
4. Discussion
4.1. Population Structure of Borrelia lusitaniae
4.2. Host and Vector Associations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kurtenbach, K.; Hoen, A.G.; Bent, S.J.; Vollmer, S.A.; Ogden, N.H.; Margos, G. Population biology of lyme borreliosis spirochetes. In Bacterial Population Genetics in Infectious Disease, 1st ed.; Robinson, D.A., Falush, D., Feil, E.J., Eds.; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2010. [Google Scholar]
- Margos, G.; Vollmer, S.A.; Ogden, N.H.; Fish, D. Population genetics, taxonomy, phylogeny and evolution of Borrelia burgdorferi sensu lato. Infect. Genet. Evol. 2011, 11, 1545–1563. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rizzoli, A.; Tagliapietra, V.; Cagnacci, F.; Marini, G.; Arnoldi, D.; Rosso, F.; Rosa, R. Parasites and wildlife in a changing world: The vector-host-pathogen interaction as a learning case. Int. J. Parasitol. Parasites Wildl. 2019, 9, 394–401. [Google Scholar] [CrossRef]
- Frank, S.A. Immunology and Evolution of Infectious Disease; Princeton University Press: Princeton, NJ, USA, 2002. [Google Scholar]
- Jongejan, F.; Uilenberg, G. The global importance of ticks. Parasitology 2004, 129, S3–S14. [Google Scholar] [CrossRef]
- Kurtenbach, K.; Hanincova, K.; Tsao, J.I.; Margos, G.; Fish, D.; Ogden, N.H. Fundamental processes in the evolutionary ecology of Lyme borreliosis. Nat. Rev. Microbiol. 2006, 4, 660–669. [Google Scholar] [CrossRef] [PubMed]
- Telford, S.R.; Goethert, H.K. Emerging tick-borne infections: Rediscovered and better characterized, or truly ‘new’? Parasitology 2004, 129, 301–327. [Google Scholar] [CrossRef] [PubMed]
- Medlock, J.M.; Hansford, K.M.; Bormane, A.; Derdakova, M.; Estrada-Peña, A.; George, J.-C.; Golovjona, I.; Jaenson, T.G.T.; Jensen, J.-K.; Jensen, P.M.; et al. Driving forces for changes in geographical distribution of Ixodes ricinus ticks in Europe. Parasites Vectors 2013, 6, 1. [Google Scholar] [CrossRef] [Green Version]
- Rizzoli, A.; Hauffe, H.C.; Carpi, G.; Vourc’h, G.I.; Neteler, M.; Rosà, R. Lyme borreliosis in Europe. Euro Surveill 2011, 16, 19906. [Google Scholar] [CrossRef]
- Becker, N.S.; Margos, G.; Blum, H.; Krebs, S.; Graf, A.; Lane, R.S.; Castillo-Ramirez, S.; Sing, A.; Fingerle, V. Recurrent evolution of host and vector association in bacteria of the Borrelia burgdorferi sensu lato species complex. BMC Genom. 2016, 17, 734. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Margos, G.; Fingerle, V.; Reynolds, S.E. Borrelia bavariensis: Vector Switch, Niche Invasion, and Geographical Spread of a Tick-Borne Bacterial Parasite. Front. Ecol. Evol. 2019, 7. [Google Scholar] [CrossRef] [Green Version]
- Vollmer, S.A.; Bormane, A.; Dinnis, R.E.; Seelig, F.; Dobson, A.D.; Aanensen, D.M.; James, M.C.; Donaghy, M.; Randolph, S.E.; Feil, E.J.; et al. Host migration impacts on the phylogeography of Lyme Borreliosis spirochaete species in Europe. Environ. Microbiol. 2011, 13, 184–192. [Google Scholar] [CrossRef]
- Vollmer, S.A.; Feil, E.J.; Chu, C.Y.; Raper, S.L.; Cao, W.C.; Kurtenbach, K.; Margos, G. Spatial spread and demographic expansion of Lyme borreliosis spirochaetes in Eurasia. Infect. Genet. Evol. 2013, 14, 147–155. [Google Scholar] [CrossRef] [PubMed]
- Ogden, N.H.; Mechai, S.; Margos, G. Changing geographic ranges of ticks and tick-borne pathogens: Drivers, mechanisms and consequences for pathogen diversity. Front. Cell. Infect. Microbiol. 2013, 3, 46. [Google Scholar] [CrossRef] [Green Version]
- Tsao, J.I. Reviewing molecular adaptations of Lyme borreliosis spirochetes in the context of reproductive fitness in natural transmission cycles. Vet. Res. 2009, 40, 36. [Google Scholar] [CrossRef] [Green Version]
- Hanincova, K.; Kurtenbach, K.; Diuk-Wasser, M.; Brei, B.; Fish, D. Epidemic spread of Lyme borreliosis, northeastern United States. Emerg. Infect. Dis. 2006, 12, 604–611. [Google Scholar] [CrossRef] [PubMed]
- Eisen, L. Vector competence studies with hard ticks and Borrelia burgdorferi sensu lato spirochetes: A review. Ticks Tick Borne Dis. 2020, 11, 101359. [Google Scholar] [CrossRef] [PubMed]
- Diuk-Wasser, M.A.; Gatewood, A.G.; Cortinas, M.R.; Yaremych-Hamer, S.; Tsao, J.; Kitron, U.; Hickling, G.; Brownstein, J.S.; Walker, E.; Piesman, J.; et al. Spatiotemporal patterns of host-seeking Ixodes scapularis nymphs (Acari: Ixodidae) in the United States. J. Med. Entomol. 2006, 43, 166–176. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mechai, S.; Margos, G.; Feil, E.J.; Lindsay, L.R.; Ogden, N.H. Complex population structure of Borrelia burgdorferi in southeastern and south central Canada as revealed by phylogeographic analysis. Appl. Environ. Microbiol. 2015, 81, 1309–1318. [Google Scholar] [CrossRef] [Green Version]
- Ogden, N.H.; Lindsay, L.R.; Hanincova, K.; Barker, I.K.; Bigras-Poulin, M.; Charron, D.F.; Heagy, A.; Francis, C.M.; O’Callaghan, C.J.; Schwartz, I.; et al. Role of migratory birds in introduction and range expansion of Ixodes scapularis ticks and of Borrelia burgdorferi and Anaplasma phagocytophilum in Canada. Appl. Environ. Microbiol. 2008, 74, 1780–1790. [Google Scholar] [CrossRef] [Green Version]
- Ogden, N.H.; Margos, G.; Aanensen, D.M.; Drebot, M.A.; Feil, E.J.; Hanincova, K.; Schwartz, I.; Tyler, S.; Lindsay, L.R. Investigation of genotypes of Borrelia burgdorferi in Ixodes scapularis ticks collected during surveillance in Canada. Appl. Environ. Microbiol. 2011, 77, 3244–3254. [Google Scholar] [CrossRef] [Green Version]
- Lane, R.S.; Loye, J.E. Lyme disease in California: Interrelationship of ixodid ticks (Acari), rodents, and Borrelia burgdorferi. J. Med. Entomol. 1991, 28, 719–725. [Google Scholar] [CrossRef]
- Girard, Y.A.; Travinsky, B.; Schotthoefer, A.; Fedorova, N.; Eisen, R.J.; Eisen, L.; Barbour, A.G.; Lane, R.S. Population structure of the lyme borreliosis spirochete Borrelia burgdorferi in the western black-legged tick (Ixodes pacificus) in Northern California. Appl. Environ. Microbiol. 2009, 75, 7243–7252. [Google Scholar] [CrossRef] [Green Version]
- Tyler, S.; Tyson, S.; Dibernardo, A.; Drebot, M.; Feil, E.J.; Graham, M.; Knox, N.C.; Lindsay, L.R.; Margos, G.; Mechai, S.; et al. Whole genome sequencing and phylogenetic analysis of strains of the agent of Lyme disease Borrelia burgdorferi from Canadian emergence zones. Sci. Rep. 2018, 8, 10552. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Walter, K.S.; Carpi, G.; Caccone, A.; Diuk-Wasser, M.A. Genomic insights into the ancient spread of Lyme disease across North America. Nat. Ecol. Evol. 2017, 1, 1569–1576. [Google Scholar] [CrossRef] [PubMed]
- Margos, G.; Tsao, J.I.; Castillo-Ramirez, S.; Girard, Y.A.; Hamer, S.A.; Hoen, A.G.; Lane, R.S.; Raper, S.L.; Ogden, N.H. Two boundaries separate Borrelia burgdorferi populations in North America. Appl. Environ. Microbiol. 2012, 78, 6059–6067. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Castillo-Ramirez, S.; Fingerle, V.; Jungnick, S.; Straubinger, R.K.; Krebs, S.; Blum, H.; Meinel, D.M.; Hofmann, H.; Guertler, P.; Sing, A.; et al. Trans-Atlantic exchanges have shaped the population structure of the Lyme disease agent Borrelia burgdorferi sensu stricto. Sci. Rep. 2016, 6, 22794. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gomez-Diaz, E.; Boulinier, T.; Sertour, N.; Cornet, M.; Ferquel, E.; McCoy, K.D. Genetic structure of marine Borrelia garinii and population admixture with the terrestrial cycle of Lyme borreliosis. Environ. Microbiol. 2011, 13, 2453–2467. [Google Scholar] [CrossRef] [PubMed]
- Comstedt, P.; Jakobsson, T.; Bergstrom, S. Global ecology and epidemiology of Borrelia garinii spirochetes. Infect. Ecol. Epidemiol. 2011, 1. [Google Scholar] [CrossRef]
- Munro, H.J.; Ogden, N.H.; Mechai, S.; Lindsay, L.R.; Robertson, G.J.; Whitney, H.; Lang, A.S. Genetic diversity of Borrelia garinii from Ixodes uriae collected in seabird colonies of the northwestern Atlantic Ocean. Ticks Tick Borne Dis. 2019, 10, 101255. [Google Scholar] [CrossRef]
- Norte, A.C.; Margos, G.; Becker, N.S.; Albino Ramos, J.; Nuncio, M.S.; Fingerle, V.; Araujo, P.M.; Adamik, P.; Alivizatos, H.; Barba, E.; et al. Host dispersal shapes the population structure of a tick-borne bacterial pathogen. Mol. Ecol. 2020, 29, 485–501. [Google Scholar] [CrossRef]
- Mtierova, Z.; Derdakova, M.; Chvostac, M.; Didyk, Y.M.; Mangova, B.; Rusnakova Taragelova, V.; Selyemova, D.; Sujanova, A.; Vaclav, R. Local Population Structure and Seasonal Variability of Borrelia garinii Genotypes in Ixodes ricinus Ticks, Slovakia. Int. J. Environ. Res. Public Health 2020, 17, 3607. [Google Scholar] [CrossRef]
- Vitorino, L.R.; Margos, G.; Feil, E.J.; Collares-Pereira, M.; Ze-Ze, L.; Kurtenbach, K. Fine-scale Phylogeographic Structure of Borrelia lusitaniae Revealed by Multilocus Sequence Typing. PLoS ONE 2008, 3, e4002. [Google Scholar] [CrossRef] [Green Version]
- Núncio, M.S.; Péter, O.; Alves, M.J.; Bacellar, F.; Filipe, A.R. Isolamento e caracterização de borrélias de Ixodes ricinus L. em Portugal. Revista Portuguesa Doenças Infecciosas 1993, 16, 175–179. [Google Scholar]
- Le Fleche, A.; Postic, D.; Girardet, K.; Peter, O.; Baranton, G. Characterization of Borrelia lusitaniae sp. nov. by 16S ribosomal DNA sequence analysis. Int. J. Syst. Bacteriol. 1997, 47, 921–925. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dsouli, N.; Younsi-Kabachii, H.; Postic, D.; Nouira, S.; Gern, L.; Bouattour, A. Reservoir role of lizard Psammodromus algirus in transmission cycle of Borrelia burgdorferi sensu lato (Spirochaetaceae) in Tunisia. J. Med. Entomol. 2006, 43, 737–742. [Google Scholar] [CrossRef]
- Amore, G.; Tomassone, L.; Grego, E.; Ragagli, C.; Bertolotti, L.; Nebbia, P.; Rosati, S.; Mannelli, A. Borrelia lusitaniae in immature Ixodes ricinus (Acari: Ixodidae) feeding on common wall lizards in Tuscany, central Italy. J. Med. Entomol. 2007, 44, 303–307. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Norte, A.C.; Alves da Silva, A.; Alves, J.; da Silva, L.P.; Nuncio, M.S.; Escudero, R.; Anda, P.; Ramos, J.A.; Lopes de Carvalho, I. The importance of lizards and small mammals as reservoirs for Borrelia lusitaniae in Portugal. Environ. Microbiol. Rep. 2015, 7, 188–193. [Google Scholar] [CrossRef] [PubMed]
- Baptista, S.; Quaresma, A.; Aires, T.; Kurtenbach, K.; Santos-Reis, M.; Nicholson, M.; Collares-Pereira, M. Lyme borreliosis spirochetes in questing ticks from mainland Portugal. Int. J. Med. Microbiol. 2004, 293 (Suppl. 37), 109–116. [Google Scholar] [CrossRef]
- Norte, A.C.; Ramos, J.A.; Gern, L.; Nuncio, M.S.; Lopes de Carvalho, I. Birds as reservoirs for Borrelia burgdorferi s.l. in Western Europe: Circulation of B. turdi and other genospecies in bird-tick cycles in Portugal. Environ. Microbiol. 2013, 15, 386–397. [Google Scholar] [CrossRef]
- De Carvalho, I.L.; Milhano, N.; Santos, A.S.; Almeida, V.; Barros, S.C.; De Sousa, R.; Nuncio, M.S. Detection of Borrelia lusitaniae, Rickettsia sp. IRS3, Rickettsia monacensis, and Anaplasma phagocytophilum in Ixodes ricinus collected in Madeira Island, Portugal. Vector Borne Zoonotic Dis. 2008, 8, 575–579. [Google Scholar] [CrossRef] [Green Version]
- Younsi, H.; Postic, D.; Baranton, G.; Bouattour, A. High prevalence of Borrelia lusitaniae in Ixodes ricinus ticks in Tunisia. Eur. J. Epidemiol. 2001, 17, 53–56. [Google Scholar] [CrossRef] [PubMed]
- Richter, D.; Matuschka, F.R. Perpetuation of the Lyme disease spirochete Borrelia lusitaniae by lizards. Appl. Environ. Microbiol. 2006, 72, 4627–4632. [Google Scholar] [CrossRef] [Green Version]
- Ragagli, C.; Bertolotti, L.; Giacobini, M.; Mannelli, A.; Bisanzio, D.; Amore, G.; Tomassone, L. Transmission dynamics of Borrelia lusitaniae and Borrelia afzelii among Ixodes ricinus, lizards, and mice in Tuscany, central Italy. Vector Borne Zoonotic Dis. 2011, 11, 21–28. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Sousa, R.; Lopes de Carvalho, I.; Santos, A.S.; Bernardes, C.; Milhano, N.; Jesus, J.; Menezes, D.; Nuncio, M.S. Role of the lizard Teira dugesii as a potential host for Ixodes ricinus tick-borne pathogens. Appl. Environ. Microbiol. 2012, 78, 3767–3769. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Majlathova, V.; Majlath, I.; Derdakova, M.; Vichova, B.; Pet’ko, B. Borrelia lusitaniae and green lizards (Lacerta viridis), Karst Region, Slovakia. Emerg. Infect. Dis. 2006, 12, 1895–1901. [Google Scholar] [CrossRef] [PubMed]
- De Carvalho, I.L.; Zeidner, N.; Ullmann, A.; Hojgaard, A.; Amaro, F.; Ze-Ze, L.; Alves, M.J.; de Sousa, R.; Piesman, J.; Nuncio, M.S. Molecular characterization of a new isolate of Borrelia lusitaniae derived from Apodemus sylvaticus in Portugal. Vector Borne Zoonotic Dis. 2010, 10, 531–534. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sarih, M.; Jouda, F.; Gern, L.; Postic, D. First isolation of Borrelia burgdorferi sensu lato from Ixodes ricinus ticks in Morocco. Vector Borne Zoonotic Dis. 2003, 3, 133–139. [Google Scholar] [CrossRef] [PubMed]
- Zhioua, E.; Bouattour, A.; Hu, C.M.; Gharbi, M.; Aeschliman, A.; Ginsberg, H.S.; Gern, L. Infection of Ixodes ricinus (Acari: Ixodidae) by Borrelia burgdorferi sensu lato in North Africa. J. Med. Entomol. 1999, 36, 216–218. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bertolotti, L.; Tomassone, L.; Tramuta, C.; Grego, E.; Amore, G.; Ambrogi, C.; Nebbia, P.; Mannelli, A. Borrelia lusitaniae and spotted fever group rickettsiae in Ixodes ricinus (Acari: Ixodidae) in Tuscany, central Italy. J. Med. Entomol. 2006, 43, 159–165. [Google Scholar] [CrossRef] [Green Version]
- Taragelova, V.R.; Mahrikova, L.; Selyemova, D.; Vaclav, R.; Derdakova, M. Natural foci of Borrelia lusitaniae in a mountain region of Central Europe. Ticks Tick Borne Dis. 2016, 7, 350–356. [Google Scholar] [CrossRef] [PubMed]
- Wodecka, B.; Skotarczak, B. First isolation of Borrelia lusitaniae DNA from Ixodes ricinus ticks in Poland. Scand. J. Infect. Dis. 2005, 37, 27–34. [Google Scholar] [CrossRef] [PubMed]
- Okeyo, M.; Hepner, S.; Rollins, R.E.; Hartberger, C.; Straubinger, R.K.; Marosevic, D.; Bannister, S.A.; Bormane, A.; Donaghy, M.; Sing, A.; et al. Longitudinal study of prevalence and spatio-temporal distribution of Borrelia burgdorferi sensu lato in ticks from three defined habitats in Latvia, 1999–2010. Environ. Microbiol. 2020. [Google Scholar] [CrossRef]
- De Michelis, S.; Sewell, H.S.; Collares-Pereira, M.; Santos-Reis, M.; Schouls, L.M.; Benes, V.; Holmes, E.C.; Kurtenbach, K. Genetic diversity of Borrelia burgdorferi sensu lato in ticks from mainland Portugal. J. Clin. Microbiol. 2000, 38, 2128–2133. [Google Scholar] [CrossRef] [PubMed]
- Zeidner, N.S.; Schneider, B.S.; Nuncio, M.S.; Gern, L.; Piesman, J. Coinoculation of Borrelia spp. with tick salivary gland lysate enhances spirochete load in mice and is tick species-specific. J. Parasitol. 2002, 88, 1276–1278. [Google Scholar] [PubMed]
- Collares-Pereira, M.; Couceiro, S.; Franca, I.; Kurtenbach, K.; Schafer, S.M.; Vitorino, L.; Goncalves, L.; Baptista, S.; Vieira, M.L.; Cunha, C. First isolation of Borrelia lusitaniae from a human patient. J. Clin. Microbiol. 2004, 42, 1316–1318. [Google Scholar] [CrossRef] [Green Version]
- Da Franca, I.; Santos, L.; Mesquita, T.; Collares-Pereira, M.; Baptista, S.; Vieira, L.; Viana, I.; Vale, E.; Prates, C. Lyme borreliosis in Portugal caused by Borrelia lusitaniae? Clinical report on the first patient with a positive skin isolate. Wiener Klinische Wochenschrift 2005, 117, 429–432. [Google Scholar] [CrossRef] [PubMed]
- Lopes de Carvalho, I.L.; Fonseca, J.E.; Marques, J.G.; Ullmann, A.; Hojgaard, A.; Zeidner, N.; Nuncio, M.S. Vasculitis-like syndrome associated with Borrelia lusitaniae infection. Clin. Rheumatol. 2008, 27, 1587–1591. [Google Scholar] [CrossRef] [PubMed]
- Grego, E.; Bertolotti, L.; Peletto, S.; Amore, G.; Tomassone, L.; Mannelli, A. Borrelia lusitaniae OspA gene heterogeneity in Mediterranean basin area. J. Mol. Evol. 2007, 65, 512–518. [Google Scholar] [CrossRef]
- Estrada-Peña, A.; Nava, S.; Petney, T. Description of all the stages of Ixodes inopinatus n. sp. (Acari: Ixodidae). Ticks Tick Borne Dis. 2014, 5, 734–743. [Google Scholar] [CrossRef]
- Noureddine, R.; Chauvin, A.; Plantard, O. Lack of genetic structure among Eurasian populations of the tick Ixodes ricinus contrasts with marked divergence from north-African populations. Int. J. Parasitol. 2010, 41, 183–192. [Google Scholar] [CrossRef]
- Falco, R.C.; Fish, D. A comparison of methods for sampling the deer tick, Ixodes dammini, in a Lyme disease endemic area. Exp. Appl. Acarol. 1992, 14, 165–173. [Google Scholar] [CrossRef]
- Rollins, R.E.; Mouchet, A.; Margos, G.; Fingerle, V.; Becker, N.S.; Dingemanse, N.J. Repeatable differences in exploratory behaviour predict tick infestation probability in wild great tits. Behav. Ecol. Sociobiol. 2021, 75, 48. [Google Scholar] [CrossRef]
- Estrada-Peña, A.; Bouattour, A.; Camicas, J.L.; Walker, A.R. Ticks of Domestic Animals in the Mediterranean Region—A Guide to Identification of Species; University of Zaragoza: Zaragoza, Spain, 2004. [Google Scholar]
- Guy, E.C.; Stanek, G. Detection of Borrelia burgdorferi in patients with Lyme disease by the polymerase chain reaction. J. Clin. Pathol. 1991, 44, 610–611. [Google Scholar] [CrossRef] [Green Version]
- Derdakova, M.; Beati, L.; Pet’ko, B.; Stanko, M.; Fish, D. Genetic variability within Borrelia burgdorferi sensu lato genospecies established by PCR-single-strand conformation polymorphism analysis of the rrfA-rrlB intergenic spacer in Ixodes ricinus ticks from the Czech Republic. Appl. Environ. Microbiol. 2003, 69, 509–516. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rijpkema, S.G.; Molkenboer, M.J.; Schouls, L.M.; Jongejan, F.; Schellekens, J.F. Simultaneous detection and genotyping of three genomic groups of Borrelia burgdorferi sensu lato in Dutch Ixodes ricinus ticks by characterization of the amplified intergenic spacer region between 5S and 23S rRNA genes. J. Clin. Microbiol. 1995, 33, 3091–3095. [Google Scholar] [CrossRef] [Green Version]
- Johnson, B.J.; Happ, C.M.; Mayer, L.W.; Piesman, J. Detection of Borrelia burgdorferi in ticks by species-specific amplification of the flagellin gene. Am. J. Trop. Med. Hyg. 1992, 47, 730–741. [Google Scholar] [CrossRef] [PubMed]
- Hidri, N.; Barraud, O.; de Martino, S.; Garnier, F.; Paraf, F.; Martin, C.; Sekkal, S.; Laskar, M.; Jaulhac, B.; Ploy, M.C. Lyme endocarditis. Clin. Microbiol. Infect. 2012, 18, E531–E532. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Margos, G.; Gatewood, A.G.; Aanensen, D.M.; Hanincova, K.; Terekhova, D.; Vollmer, S.A.; Cornet, M.; Piesman, J.; Donaghy, M.; Bormane, A.; et al. MLST of housekeeping genes captures geographic population structure and suggests a European origin of Borrelia burgdorferi. Proc. Natl. Acad. Sci. USA 2008, 105, 8730–8735. [Google Scholar] [CrossRef] [Green Version]
- Margos, G.; Vollmer, S.A.; Cornet, M.; Garnier, M.; Fingerle, V.; Wilske, B.; Bormane, A.; Vitorino, L.; Collares-Pereira, M.; Drancourt, M.; et al. A new Borrelia species defined by Multilocus Sequence Analysis of Housekeeping Genes. Appl. Environ. Microbiol. 2009, 75, 5410–5416. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mangold, A.J.; Bargues, M.D.; Mas-Coma, S. Mitochondrial 16S rDNA sequences and phylogenetic relationships of species of Rhipicephalus and other tick genera among Metastriata (Acari: Ixodidae). Parasitol. Res. 1998, 84, 478–484. [Google Scholar] [CrossRef]
- Huson, D.H.; Bryant, D. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 2006, 23, 254–267. [Google Scholar] [CrossRef]
- Croucher, N.J.; Page, A.J.; Connor, T.R.; Delaney, A.J.; Keane, J.A.; Bentley, S.D.; Parkhill, J.; Harris, S.R. Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res. 2015, 43, e15. [Google Scholar] [CrossRef] [Green Version]
- Bruen, T.C.; Philippe, H.; Bryant, D. A simple and robust statistical test for detecting the presence of recombination. Genetics 2006, 172, 2665–2681. [Google Scholar] [CrossRef] [Green Version]
- Grana-Miraglia, L.; Evans, B.A.; Lopez-Jacome, L.E.; Hernandez-Duran, M.; Colin-Castro, C.A.; Volkow-Fernandez, P.; Cevallos, M.A.; Franco-Cendejas, R.; Castillo-Ramirez, S. Origin of OXA-23 Variant OXA-239 from a Recently Emerged Lineage of Acinetobacter baumannii International Clone V. mSphere 2020, 5. [Google Scholar] [CrossRef] [PubMed]
- Cheng, L.; Connor, T.R.; Sirén, J.; Aanensen, D.M.; Corander, J. Hierarchical and spatially explicit clustering of DNA sequences with BAPS software. Mol. Biol. Evol. 2013, 30, 1224–1228. [Google Scholar] [CrossRef]
- Stecher, G.; Tamura, K.; Kumar, S. Molecular Evolutionary Genetics Analysis (MEGA) for macOS. Mol. Biol. Evol. 2020, 37, 1237–1239. [Google Scholar] [CrossRef] [PubMed]
- James, G.; Witten, D.; Hastie, T.; Tibshirani, R. An Introduction to Statistical Learning, 1st ed.; Springer: New York, NY, USA, 2013. [Google Scholar] [CrossRef]
- Gower, J.C. A general coefficient of similarity and some of its properties. Biometrics 1971, 27, 857–874. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2014. [Google Scholar]
- Galili, T. Dendextend: An R package for visualizing, adjusting and comparing trees of hierarchical clustering. Bioinformatics 2015, 31, 3718–3720. [Google Scholar] [CrossRef] [Green Version]
- Francisco, A.P.; Vaz, C.; Monteiro, P.T.; Melo-Cristino, J.; Ramirez, M.; Carrico, J.A. PHYLOViZ: Phylogenetic inference and data visualization for sequence based typing methods. BMC Bioinform. 2012, 13, 87. [Google Scholar] [CrossRef] [Green Version]
- Francisco, A.P.; Bugalho, M.; Ramirez, M.; Carrico, J.A. Global optimal eBURST analysis of multilocus typing data using a graphic matroid approach. BMC Bioinform. 2009, 10, 152. [Google Scholar] [CrossRef] [Green Version]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. jModelTest 2: More models, new heuristics and parallel computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef] [Green Version]
- Guindon, S.; Gascuel, O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 2003, 52, 696–704. [Google Scholar] [CrossRef] [Green Version]
- Pérez-Eid, C. Les Tiques: Identification, Biologie, Importance Médicale et Veterinaire; Lavoisier: Paris, France, 2007. [Google Scholar]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Jacquot, M.; Gonnet, M.; Ferquel, E.; Abrial, D.; Claude, A.; Gasqui, P.; Choumet, V.; Charras-Garrido, M.; Garnier, M.; Faure, B.; et al. Comparative population genomics of the Borrelia burgdorferi species complex reveals high degree of genetic isolation among species and underscores benefits and constraints to studying intra-specific epidemiological processes. PLoS ONE 2014, 9, e94384. [Google Scholar] [CrossRef] [PubMed]
- Shapiro, B.J.; Friedman, J.; Cordero, O.X.; Preheim, S.P.; Timberlake, S.C.; Szabo, G.; Polz, M.F.; Alm, E.J. Population genomics of early events in the ecological differentiation of bacteria. Science 2012, 336, 48–51. [Google Scholar] [CrossRef] [Green Version]
- Shapiro, B.J. Signatures of natural selection and ecological differentiation in microbial genomes. Adv. Exp. Med. Biol. 2014, 781, 339–359. [Google Scholar] [CrossRef] [PubMed]
- Frey, W.; Lösch, R. Geobotanik—Pflanze und Vegetation in Raum und Zeit; Spektrum Akademischer Verlag: Munich, Germany; Heidelberg, Germany, 2010. [Google Scholar]
- Poli, P.; Lenoir, J.; Plantard, O.; Ehrmann, S.; Roed, K.H.; Leinaas, H.P.; Panning, M.; Guiller, A. Strong genetic structure among populations of the tick Ixodes ricinus across its range. Ticks Tick Borne Dis. 2020, 11, 101509. [Google Scholar] [CrossRef] [PubMed]
- Carranza, S.; Harris, D.J.; Arnold, E.N.; Batista, V.; Gonzalez de la Vega, J.P. Phylogeography of the lacertid lizard, Psammodromus algirus, in Iberia and across the Strait of Gibraltar. J. Biogeogr. 2006, 33, 1279–1288. [Google Scholar] [CrossRef]
- Verdú Ricoy, J.; Carranza, S.; Salvador, A.; Busack, S.; Díaz, J. Phylogeography of Psammodromus algirus (Lacertidae) revisited: Systematic implications. Amphib. Reptil. 2010, 31, 576–582. [Google Scholar]
- Younsi, H.; Sarih, M.; Jouda, F.; Godfroid, E.; Gern, L.; Bouattour, A.; Baranton, G.; Postic, D. Characterization of Borrelia lusitaniae isolates collected in Tunisia and Morocco. J. Clin. Microbiol. 2005, 43, 1587–1593. [Google Scholar] [CrossRef] [Green Version]




| Tick AN for NCBI_16S rRNA | Tick AN for NCBI_trospA | Tick Species (16S rRNA Based ID) | Year | Stage/Sex | B. lusitaniae Infection Status | MLST B. lusitaniae | Region Specific † | Reference | |
|---|---|---|---|---|---|---|---|---|---|
| GU074596TN | GU074839TN | Ixodes inopinatus # | F | unknown | TN, Jbel el Jouza | [61] | |||
| GU074598DZ ‡ | GU074841DZ | Ixodes inopinatus # | F | unknown | DZ, El Tarf | [61] | |||
| GU074602MA | GU074845MA | Ixodes inopinatus # | M | unknown | MA, Taza | [61] | |||
| GU074597FR | GU074840FR ‡ | Ixodes ricinus | F | unknown | FR, Forêt de Chizé | [61] | |||
| GU074606ES ‡ | GU074849ES | Ixodes ricinus | F | unknown | ES, Otxandio | [61] | |||
| GU074595IE ‡ | GU074838IE | Ixodes ricinus | F | unknown | IE, Cork, Killarney National park | [61] | |||
| GU074603GB | GU074846GB | Ixodes ricinus | N | unknown | UK | [61] | |||
| GU074607NL | GU074850NL | Ixodes ricinus | F | unknown | NL | [61] | |||
| GU074605DE ‡ | GU074848DE | Ixodes ricinus | N | unknown | DE, Munich, English garden | [61] | |||
| GU074599IR | GU074842IR | Ixodes ricinus | F | unknown | IR, Mazendaran province | [61] | |||
| GU074592DK | GU074835DK | Ixodes ricinus | F | unknown | DK, Grib Skov Forest | [61] | |||
| GU074593SE ‡ | GU074836SE | Ixodes ricinus | F | unknown | SE, Alsike | [61] | |||
| GU074588SK | GU074831SK | Ixodes ricinus | F | unknown | SK, Železná Studnička | [61] | |||
| GU074589SK ‡ | GU074832SK ‡ | Ixodes ricinus | F | unknown | SK, Vel’ký Lom | [61] | |||
| GU074590SK | GU074833SK | Ixodes ricinus | F | unknown | SK, Malá Lehota | [61] | |||
| GU074594EE ‡ | GU074837EE ‡ | Ixodes ricinus | F | unknown | EE, Tartumaa | [61] | |||
| GU074600FI | GU074843FI | Ixodes ricinus | F | unknown | FI, Turku archipelago | [61] | |||
| GU074604HU | GU074847HU | Ixodes ricinus | F | unknown | HU, Mátrafüred forest | [61] | |||
| GU074591MD | GU074834MD ‡ | Ixodes ricinus | F | unknown | MD | [61] | |||
| GU074601BG | GU074844BG | Ixodes ricinus | F | unknown | BG, Sofia | [61] | |||
| 13310PT18 ‡ | 13310PT18 | Ixodes ricinus | 2018 | F | positive | south Tagus | PT, Santiago do Cacém | This study | |
| 13360PT18 ‡ | 13360PT18 | Ixodes ricinus | 2018 | F | positive | south Tagus | PT, Grândola | This study | |
| 14401PT18 ‡ | 14401PT18 | Ixodes ricinus | 2018 | F | positive | south Tagus | PT, Santiago do Cacém | This study | |
| G101PT09 ‡ | G101PT09 | Ixodes ricinus | 2009 | N | unknown | north Tagus | PT, Gerês | This study | |
| R1756PT13 ‡ | R1756PT13 | Ixodes ricinus | 2013 | N | positive | north Tagus | PT, Mafra | This study | |
| R1773PT13 ‡ | R1773PT13 | Ixodes ricinus | 2013 | N | positive | north Tagus | PT, Mafra | This study | |
| R1812PT13 ‡ | R1812PT13 | Ixodes ricinus | 2013 | N | negative | north Tagus | PT, Mafra | This study | |
| R1794PT13 | R1794PT13 | Ixodes ricinus | 2013 | N | negative | north Tagus | PT, Mafra | This study | |
| 3117DZ16 | 3117DZ16 | Ixodes inopinatus | 2016 | positive | yes: ID 3117 | DZ, Kabylie | This study | ||
| T73T04DZ16 ‡ | T73T04DZ16 | Ixodes ricinus | 2016 | positive | DZ, Kabylie | This study | |||
| 3115DZ16 | 3115DZ16 | Ixodes ricinus | 2016 | positive | DZ, Kabylie | This study | |||
| MH152SK17 ‡ | MH152SK17 | Ixodes ricinus | 2017 | M | positive | SK, Martinské hole | This study | ||
| MH149SK14 ‡ | MH149SK14 | Ixodes ricinus | 2014 | M | positive | SK, Martinské hole | This study | ||
| MH139SK14 | MH139SK14 | Ixodes ricinus | 2014 | M | positive | yes: ID 2653 | SK, Martinské hole | This study | |
| MH106SK17 ‡ | MH106SK17 | Ixodes ricinus | 2017 | F | positive | SK, Martinské hole | This study | ||
| MH60SK17 ‡ | MH60SK17 | Ixodes ricinus | 2017 | F | positive | SK, Martinské hole | This study | ||
| 2874SK16 ‡ | 2874SK16 | Ixodes ricinus | 2016 | F | positive | SK, Martinské hole | This study | ||
| MH126SK14 ‡ | MH126SK14 | Ixodes ricinus | 2014 | F | positive | SK, Martinské hole | This study | ||
| 2652SK16 | 2652SK16 | Ixodes ricinus | 2016 | F | positive | yes: ID 2652 | SK, Martinské hole | This study | |
| 11-E12 | 11-E12 | Ixodes inopinatus | 2018 | N | negative | DE, Starnberg | This study | ||
| 7-F5 | 7-F5 | Ixodes inopinatus | 2018 | N | negative | DE, Starnberg | This study | ||
| 8-C12 | 8-C12 | Ixodes inopinatus | 2018 | N | negative | DE, Starnberg | This study | ||
| Sample No. | Sample ID | Strain | Genospecies | Country | Area | Continent | ST | Year of Collection | Biological Source | Tick Code | Reference |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 59 | 162/11b | Borrelia lusitaniae | Serbia | Belgrade-Titov gaj | Europe | 628 | 2011 | Ixodes ricinus | MLST DB | |
| 2 | 63 | 167/11b | Borrelia lusitaniae | Serbia | Belgrade-Kosutnjak | Europe | 628 | 2011 | Ixodes ricinus | MLST DB | |
| 3 | 64 | 167/11c | Borrelia lusitaniae | Serbia | Belgrade-Kosutnjak | Europe | 136 | 2011 | Ixodes ricinus | MLST DB | |
| 4 | 124 | PoTiBGr41 | Borrelia lusitaniae | Portugal | Grândola | Europe | 60 | 2002 | Ixodes ricinus | MLST DB | |
| 5 | 125 | PoTiBGr82 | Borrelia lusitaniae | Portugal | Grândola | Europe | 61 | 2002 | Ixodes ricinus | MLST DB | |
| 6 | 126 | PoTiBGr130 | Borrelia lusitaniae | Portugal | Grândola | Europe | 62 | 2003 | Ixodes ricinus | MLST DB | |
| 7 | 127 | PoTiBGr131 | Borrelia lusitaniae | Portugal | Grândola | Europe | 63 | 2003 | Ixodes ricinus | MLST DB | |
| 8 | 128 | PoTiBGr136 | Borrelia lusitaniae | Portugal | Grândola | Europe | 64 | 2003 | Ixodes ricinus | MLST DB | |
| 9 | 129 | PoTibGr409 | Borrelia lusitaniae | Portugal | Grândola | Europe | 64 | 2003 | Ixodes ricinus | MLST DB | |
| 10 | 130 | PoTiBGr143 | Borrelia lusitaniae | Portugal | Grândola | Europe | 65 | 2003 | Ixodes ricinus | MLST DB | |
| 11 | 131 | PoTiBGr211 | Borrelia lusitaniae | Portugal | Grândola | Europe | 65 | 2003 | Ixodes ricinus | MLST DB | |
| 12 | 132 | PoTiBGr209 | Borrelia lusitaniae | Portugal | Grândola | Europe | 66 | 2003 | Ixodes ricinus | MLST DB | |
| 13 | 133 | PoTiBGr213 | Borrelia lusitaniae | Portugal | Grândola | Europe | 66 | 2003 | Ixodes ricinus | MLST DB | |
| 14 | 134 | PoTiBGr288 | Borrelia lusitaniae | Portugal | Grândola | Europe | 67 | 2003 | Ixodes ricinus | MLST DB | |
| 15 | 135 | PoTiBGr293 | Borrelia lusitaniae | Portugal | Grândola | Europe | 68 | 2003 | Ixodes ricinus | MLST DB | |
| 16 | 136 | PoHL1 | Borrelia lusitaniae | Portugal | Lisbon | Europe | 69 | 2002 | human | MLST DB | |
| 17 | 137 | PoTiBL37 | Borrelia lusitaniae | Portugal | Mafra | Europe | 69 | 1999 | Ixodes ricinus | MLST DB | |
| 18 | 249 | PotiB2 | Borrelia lusitaniae | Portugal | Europe | 64 | Ixodes ricinus | MLST DB | |||
| 19 | 374 | 71412L | Borrelia lusitaniae | Latvia | Europe | 218 | 2007 | Ixodes ricinus | MLST DB | ||
| 20 | 1164 | 0911001A | Borrelia lusitaniae | Austria | North-Tirol | Europe | 336 | 2009 | Ixodes ricinus | MLST DB | |
| 21 | 1181 | 0921035A | Borrelia lusitaniae | Austria | North-Tirol | Europe | 345 | 2009 | Ixodes ricinus | MLST DB | |
| 22 | 1182 | 0921036A | Borrelia lusitaniae | Austria | North-Tirol | Europe | 346 | 2009 | Ixodes ricinus | MLST DB | |
| 23 | 1250 | 233/13b | Borrelia lusitaniae | Serbia | Belgrade-Avala | Europe | 148 | 2013 | Ixodes ricinus | MLST DB | |
| 24 | 1252 | 221/10c | Borrelia lusitaniae | Serbia | Belgrade-Titov gaj | Europe | 153 | 2013 | Ixodes ricinus | MLST DB | |
| 25 | 1253 | 225/10a | Borrelia lusitaniae | Serbia | Belgrade-Titov gaj | Europe | 630 | 2013 | Ixodes ricinus | MLST DB | |
| 26 | 1254 | 228/10a | Borrelia lusitaniae | Serbia | Belgrade-Titov gaj | Europe | 194 | 2013 | Ixodes ricinus | MLST DB | |
| 27 | 1255 | 229/10a | Borrelia lusitaniae | Serbia | Belgrade-Kosutnjak | Europe | 209 | 2013 | Ixodes ricinus | MLST DB | |
| 28 | 1585 | 76/12a | Borrelia lusitaniae | Serbia | Eastern Serbia-Dobra (Djerdap Gorge) | Europe | 580 | 2012 | Ixodes ricinus | MLST DB | |
| 29 | 1586 | 77/12b | Borrelia lusitaniae | Serbia | Eastern Serbia-Dobra (Djerdap Gorge) | Europe | 586 | 2012 | Ixodes ricinus | MLST DB | |
| 30 | 1815 | 220/10b | Borrelia lusitaniae | Serbia | Belgrade-Kosutnjak | Europe | 628 | 2010 | Ixodes ricinus | MLST DB | |
| 31 | 1817 | 222/10d | Borrelia lusitaniae | Serbia | Belgrade-Titov gaj | Europe | 629 | 2010 | Ixodes ricinus | MLST DB | |
| 32 | 1819 | 224/10c | Borrelia lusitaniae | Serbia | Belgrade-Titov gaj | Europe | 630 | 2010 | Ixodes ricinus | MLST DB | |
| 33 | 1821 | 226/10d | Borrelia lusitaniae | Serbia | Belgrade-Titov gaj | Europe | 631 | 2010 | Ixodes ricinus | MLST DB | |
| 34 | 1822 | 227/10c | Borrelia lusitaniae | Serbia | Belgrade-Titov gaj | Europe | 630 | 2010 | Ixodes ricinus | MLST DB | |
| 35 | 2073 | PoTiB3 | Borrelia lusitaniae | Portugal | Europe | 766 | 1993 | Ixodes ricinus | MLST DB | ||
| 36 | 2500 | 82NCHG | Borrelia lusitaniae | Croatia | Grabovac | Europe | 900 | 2011 | Ixodes ricinus | This study | |
| 37 | 2525 | PotiBmfP147 | Borrelia lusitaniae | Portugal | Mafra | Europe | 7-011 | 2003 | Ixodes ricinus | MLST DB | |
| 38 | 2526 | PotiBmfP220 | Borrelia lusitaniae | Portugal | Mafra | Europe | 7-004 | 2003 | Ixodes ricinus | MLST DB | |
| 39 | 2527 | PotiBmfJ2 | Borrelia lusitaniae | Portugal | Mafra | Europe | 7-005 | 2001 | Ixodes ricinus | MLST DB | |
| 40 | 2528 | PotiBmfJ50 | Borrelia lusitaniae | Portugal | Mafra | Europe | 7-005 | 2003 | Ixodes ricinus | MLST DB | |
| 41 | 2529 | PotiBmfP364 | Borrelia lusitaniae | Portugal | Mafra | Europe | 7-006 | 2003 | Ixodes ricinus | MLST DB | |
| 42 | 2594 | 658 UA | Borrelia lusitaniae | Ukraine | Kyiv, M.M. Gryshko Nat. Bot. Garden | Europe | 909 | 2015 | Ixodes ricinus | This study | |
| 43 | 2652 | MH2016F8 | Borrelia lusitaniae | Slovakia | Martinské hole mountain | Europe | 918 | 2016 | Ixodes ricinus | 2652SK16 | This study |
| 44 | 2653 | SMHM139 | Borrelia lusitaniae | Slovakia | Martinské hole mountain | Europe | 919 | 2013 | Ixodes ricinus | MH139SK14 | This study |
| 52 | 3126 | PoTiB10/M436 | Borrelia lusitaniae | Portugal | Madeira | Europe | 925 | 2009 | Ixodes ricinus | This study | |
| 53 | 3127 | PoTiB11/T3087 | Borrelia lusitaniae | Portugal | Coimbra | Europe | 926 | 2014 | This study | ||
| 51 | 3125 | PoTiB9/B88 | Borrelia lusitaniae | Portugal | Santiago do Cacém | Europe | 924 | 2009 | Dermacentor marginatum | This study | |
| 45 | 3114 | Tube101-Tick25/run1-2 | Borrelia lusitaniae | Algeria | Kabylie | Africa | 7-000 | 2016 | Ixodes ricinus | This study | |
| 46 | 3117 | Tube104-Tick14/run1-5 | Borrelia lusitaniae | Algeria | Kabylie | Africa | 7-001 | 2016 | Ixodes ricinus | 3117DZ16 | This study |
| 47 | 3118 | Tube94-Tick5/run1-6 | Borrelia lusitaniae | Algeria | Kabylie | Africa | 7-002 | 2016 | Ixodes ricinus | This study | |
| 48 | 3120 | Tube101-Tick24/run1-11 | Borrelia lusitaniae | Algeria | Kabylie | Africa | 7-003 | 2016 | Ixodes ricinus | This study | |
| 49 | 3119 | Tube 95-Tick5/run1-9 | Borrelia lusitaniae | Algeria | Kabylie | Africa | 7-007 | 2016 | Ixodes ricinus | This study | |
| 50 | 3123 | Tube94-tick6/run2-7 | Borrelia lusitaniae | Algeria | Kabylie | Africa | 7-008 | 2016 | Ixodes ricinus | This study | |
| 54 | 2873 | MH2016F1 | Borrelia lusitaniae | Slovakia | Martinské hole mountain | Europe | 7-009 | 2016 | Ixodes ricinus | This study | |
| 55 | 2874 | MH2016F9 | Borrelia lusitaniae | Slovakia | Martinské hole mountain | Europe | 7-010 | 2016 | Ixodes ricinus | This study |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Norte, A.C.; Boyer, P.H.; Castillo-Ramirez, S.; Chvostáč, M.; Brahami, M.O.; Rollins, R.E.; Woudenberg, T.; Didyk, Y.M.; Derdakova, M.; Núncio, M.S.; et al. The Population Structure of Borrelia lusitaniae Is Reflected by a Population Division of Its Ixodes Vector. Microorganisms 2021, 9, 933. https://doi.org/10.3390/microorganisms9050933
Norte AC, Boyer PH, Castillo-Ramirez S, Chvostáč M, Brahami MO, Rollins RE, Woudenberg T, Didyk YM, Derdakova M, Núncio MS, et al. The Population Structure of Borrelia lusitaniae Is Reflected by a Population Division of Its Ixodes Vector. Microorganisms. 2021; 9(5):933. https://doi.org/10.3390/microorganisms9050933
Chicago/Turabian StyleNorte, Ana Cláudia, Pierre H. Boyer, Santiago Castillo-Ramirez, Michal Chvostáč, Mohand O. Brahami, Robert E. Rollins, Tom Woudenberg, Yuliya M. Didyk, Marketa Derdakova, Maria Sofia Núncio, and et al. 2021. "The Population Structure of Borrelia lusitaniae Is Reflected by a Population Division of Its Ixodes Vector" Microorganisms 9, no. 5: 933. https://doi.org/10.3390/microorganisms9050933
APA StyleNorte, A. C., Boyer, P. H., Castillo-Ramirez, S., Chvostáč, M., Brahami, M. O., Rollins, R. E., Woudenberg, T., Didyk, Y. M., Derdakova, M., Núncio, M. S., Carvalho, I. L. d., Margos, G., & Fingerle, V. (2021). The Population Structure of Borrelia lusitaniae Is Reflected by a Population Division of Its Ixodes Vector. Microorganisms, 9(5), 933. https://doi.org/10.3390/microorganisms9050933







