Unraveling a Lignocellulose-Decomposing Bacterial Consortium from Soil Associated with Dry Sugarcane Straw by Genomic-Centered Metagenomics
Abstract
1. Introduction
2. Materials and Methods
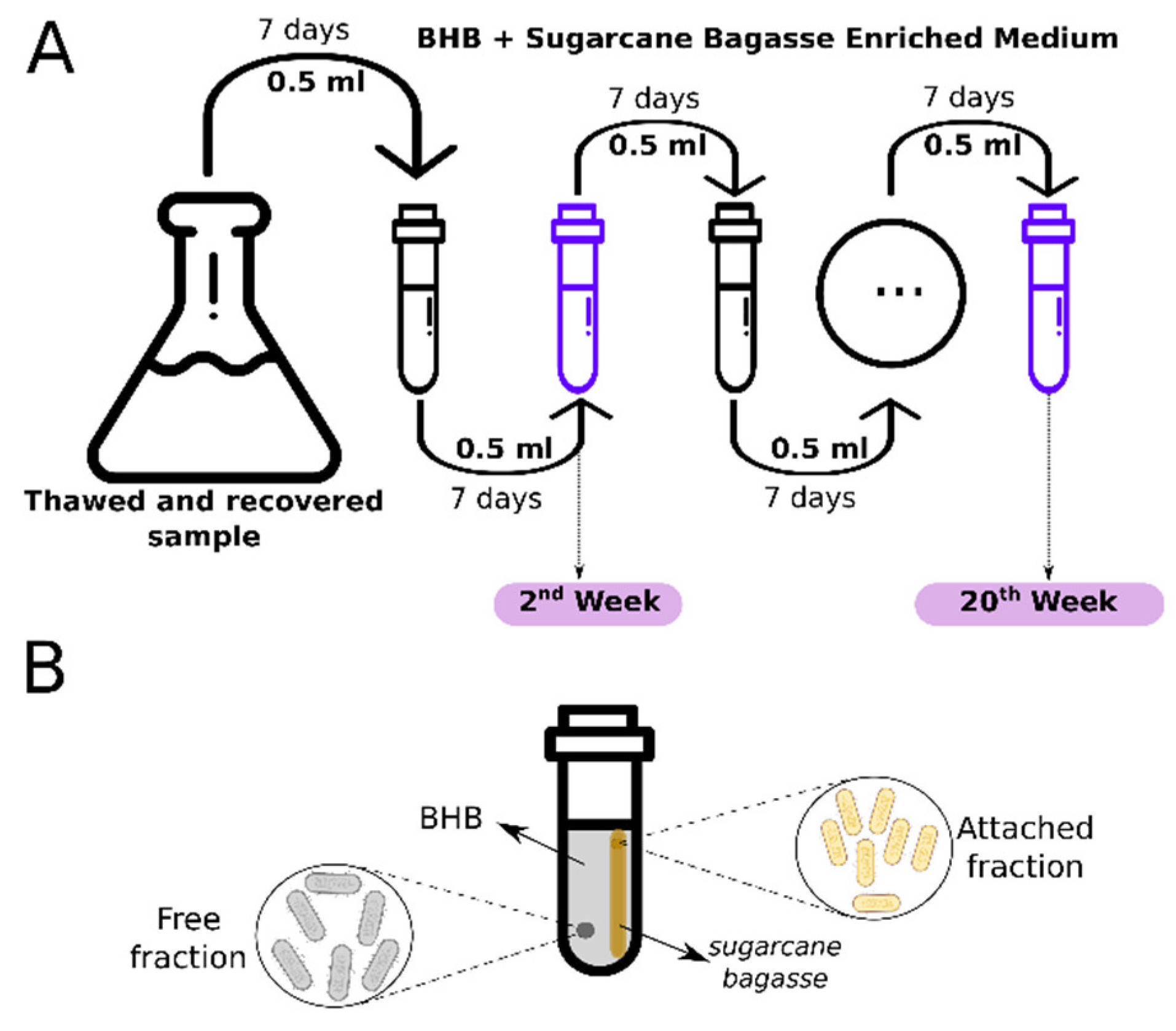
2.1. Sampling of Lignocellulose-Deconstructing Bacterial Consortium
2.2. Adaptation of the Bacterial Consortium to the Culture Medium
2.3. Metagenomic DNA Extraction and Sequencing
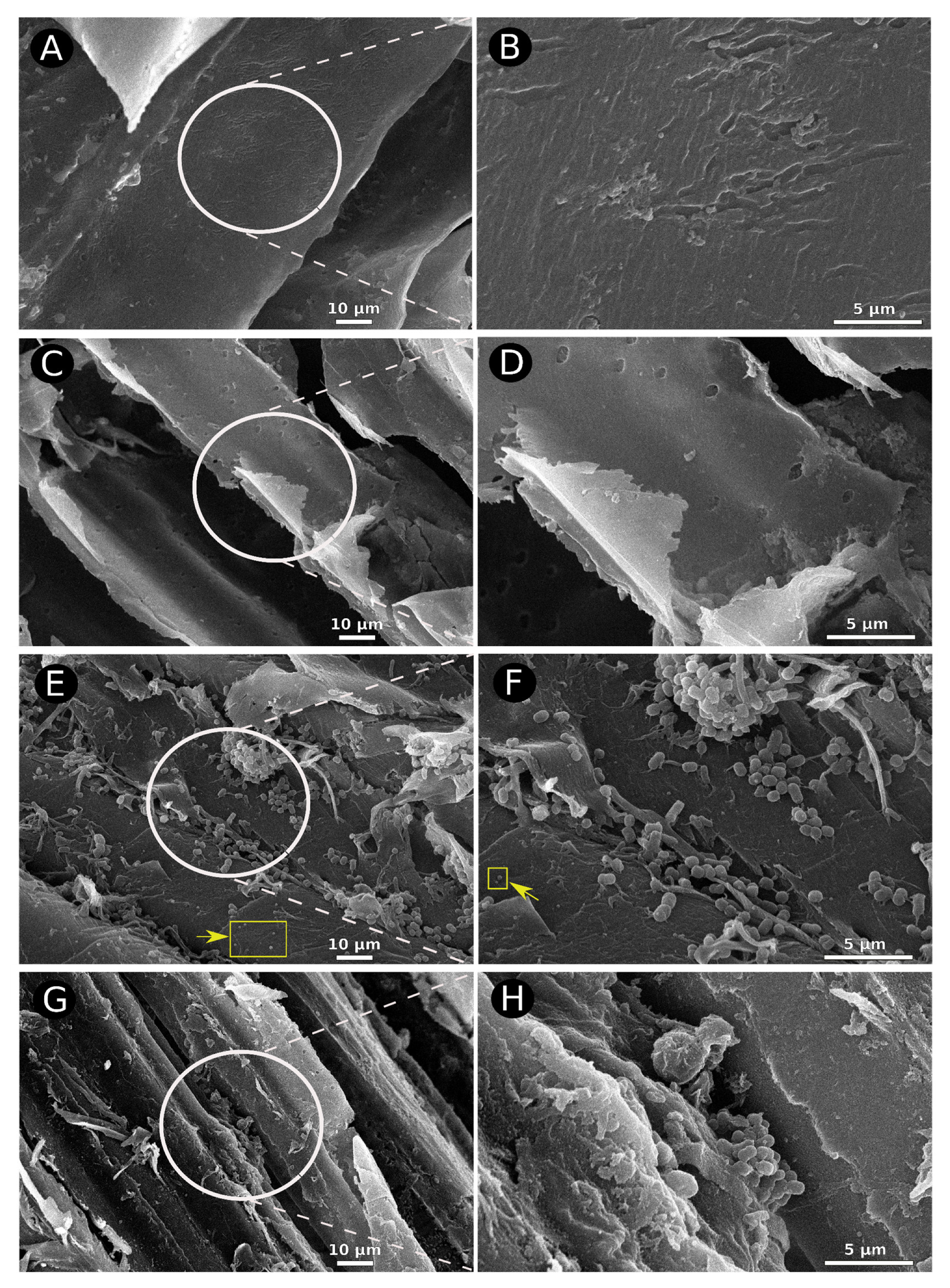
2.4. Scanning Electron Microscopy of Sugarcane Bagasse Fibers
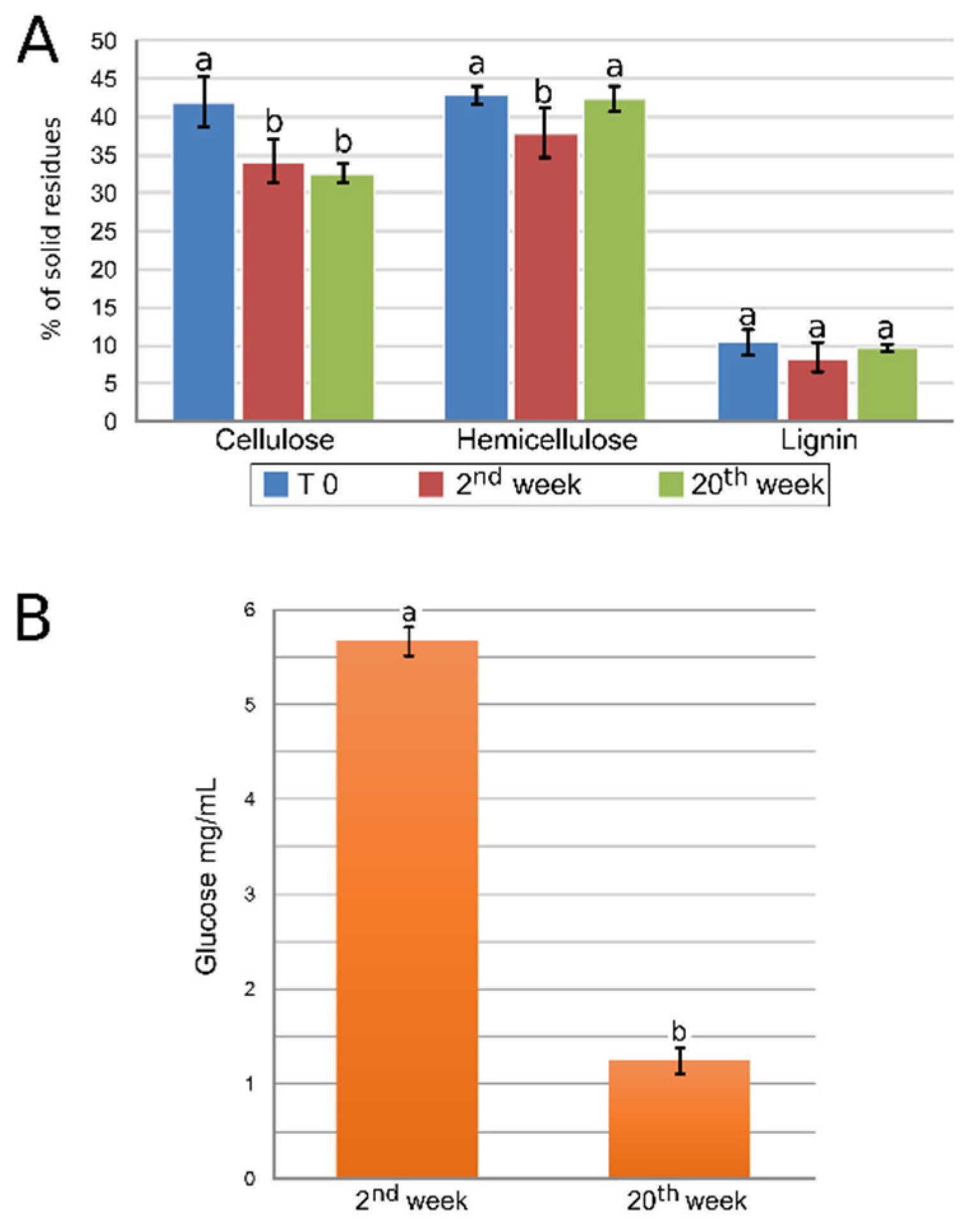
2.5. Evaluation of the Decomposition of Lignocellulosic Biomass
2.6. High-Performance Liquid Chromatography and Sugar Yields in Culture Medium
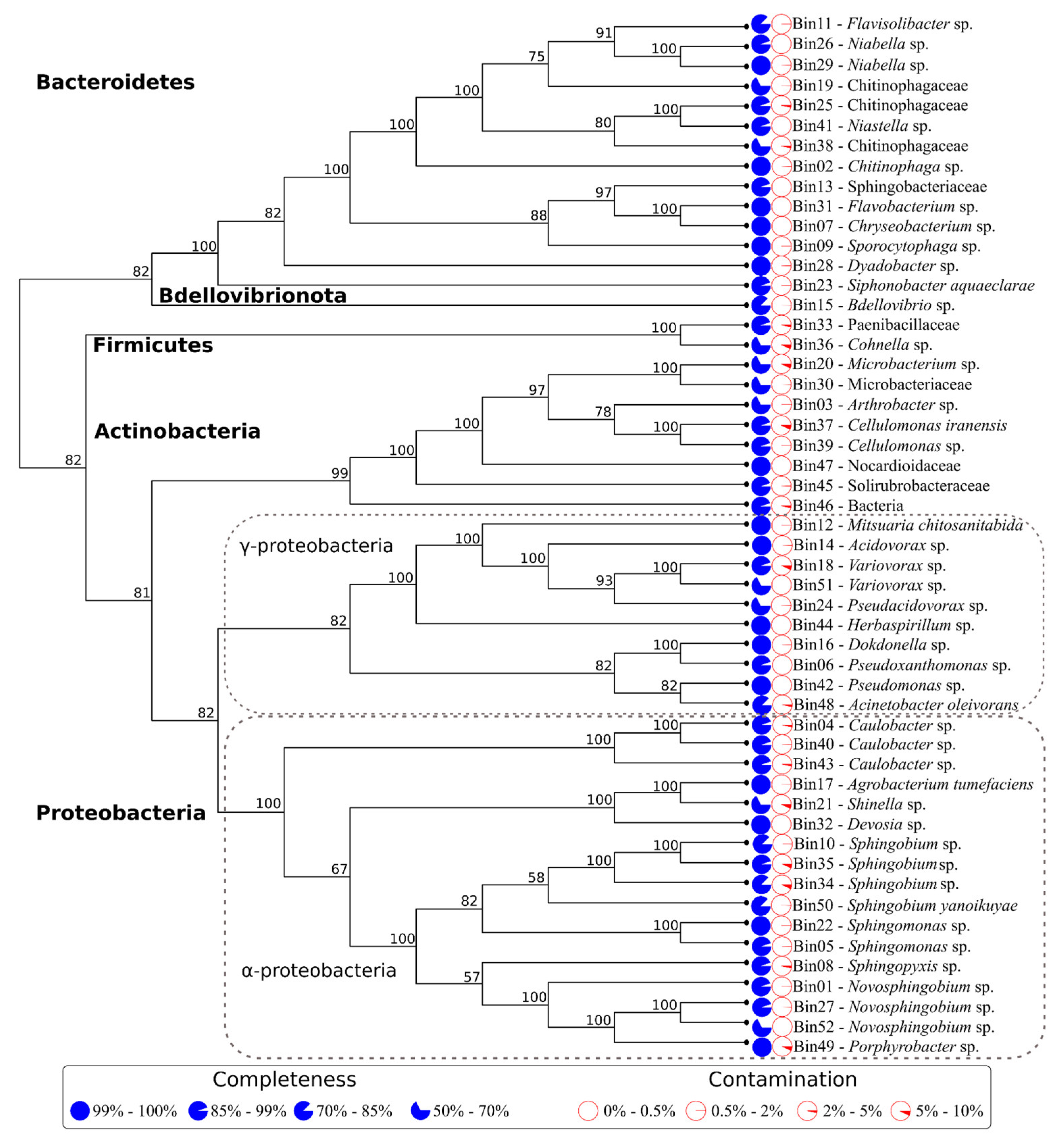
2.7. Metagenomic Binning, Quality Assessment, and Taxonomy Assignment
2.8. Functional, Metabolic Pathways and Carbohydrate Hydrolases Annotation and Analysis
2.9. Phylogenetic Analysis of the Identified MAGs
3. Results
3.1. Scanning Electron Microscopy Suggests the Role of the Consortium in the Deconstruction of Lignocellulose Biomass
3.2. Polysaccharide and Glucose Quantification Indicates a Dynamic Process of Lignocellulosic Biomass Deconstruction
3.3. Metagenome Characterization Uncovered Four Main Bacterial phyla in the Lignocellulolytic Community
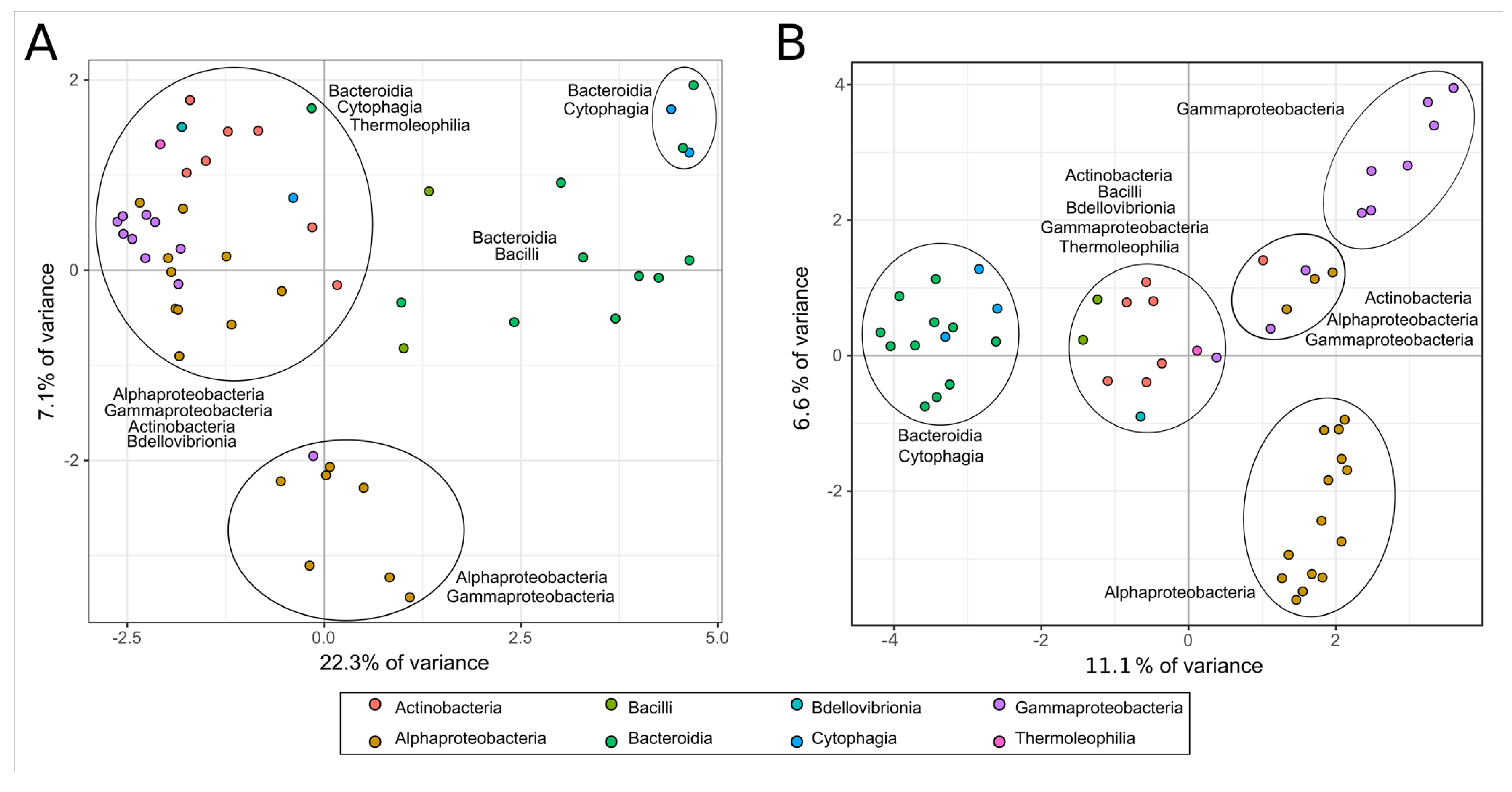
3.4. Species Relative Abundance Changes Indicate a Dynamic Community Deconstructing the Lignocellulosic Biomass
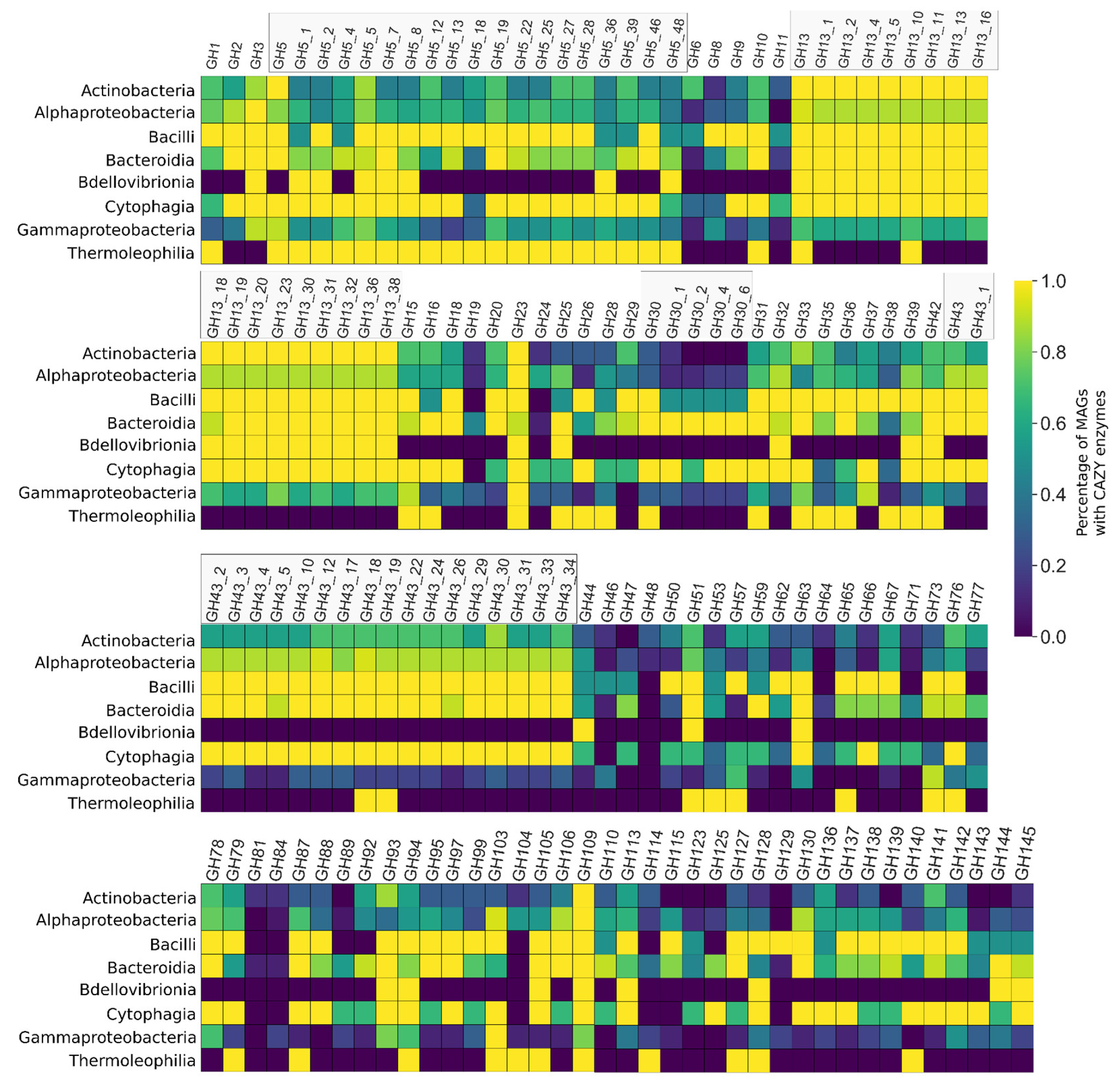
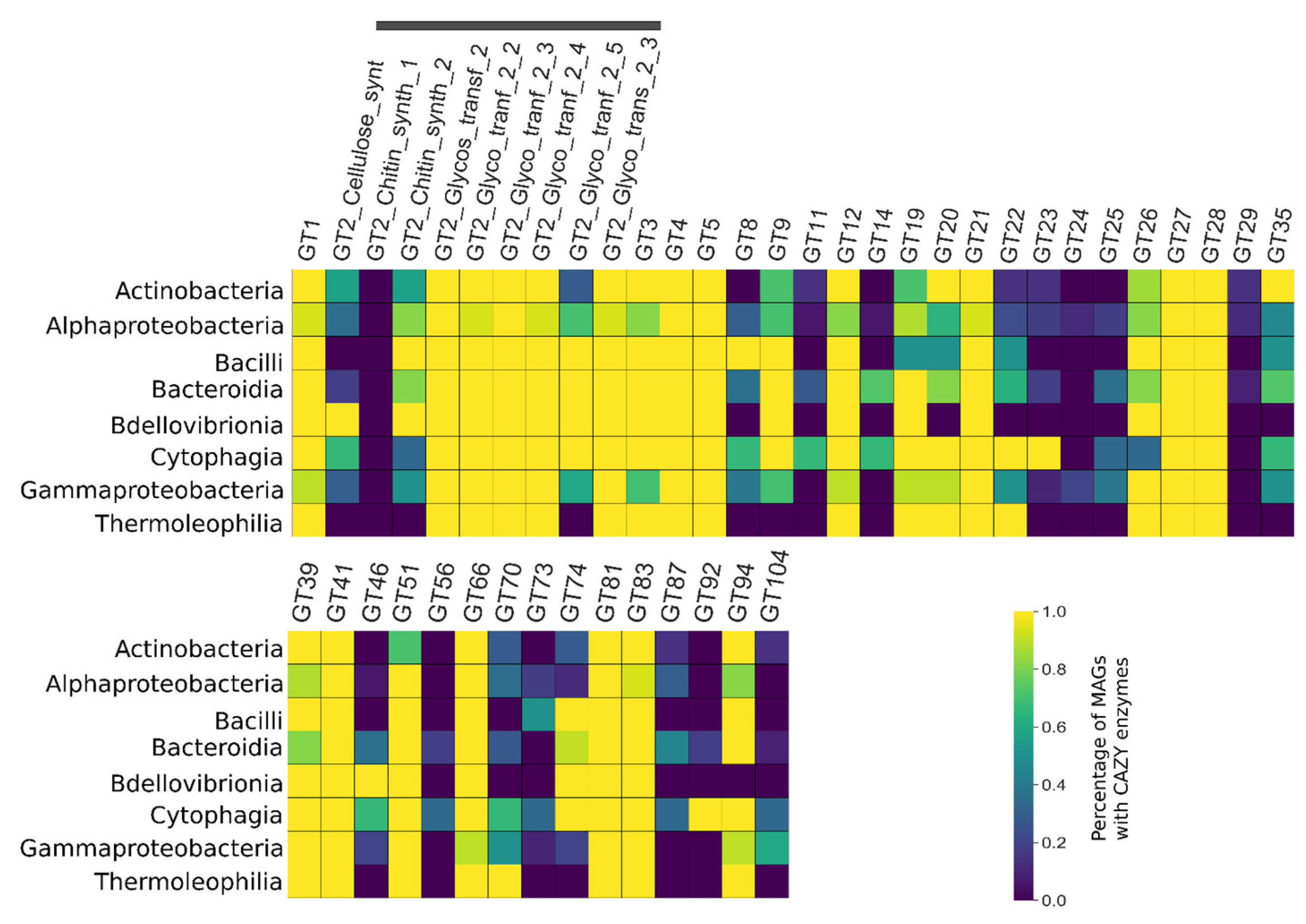
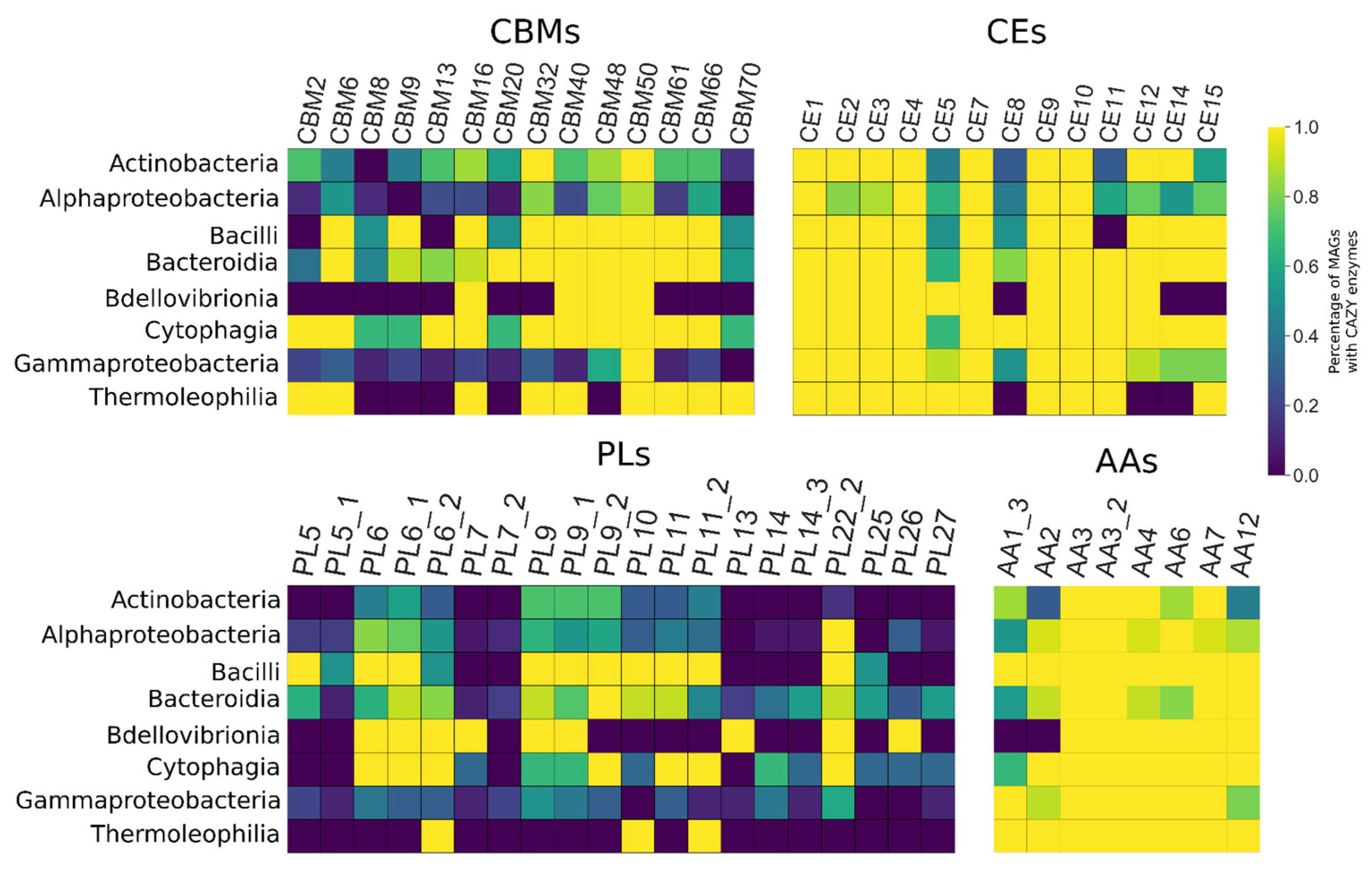
3.5. CAZY Enzymes Abundance and Distribution Indicates a Synergistic Action of Each MAG to Degrade the Lignocellulosic Mass
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Alonso Pippo, W.; Luengo, C.A.; Alonsoamador Morales Alberteris, L.; Garzone, P.; Cornacchia, G. Energy Recovery from Sugarcane-Trash in the Light of 2nd Generation Biofuels. Part 1: Current Situation and Environmental Aspects. Waste Biomass Valorization 2011, 2, 1–16. [Google Scholar] [CrossRef]
- Limayem, A.; Ricke, S.C. Lignocellulosic biomass for bioethanol production: Current perspectives, potential issues and future prospects. Prog. Energy Combust. Sci. 2012, 38, 449–467. [Google Scholar] [CrossRef]
- Bond, J.Q.; Alonso, D.M.; Dumesic, J.A. Catalytic Strategies for Converting Lignocellulosic Carbohydrates to Fuels and Chemicals. In Aqueous Pretreatment of Plant Biomass for Biological and Chemical Conversion to Fuels and Chemicals; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2013; pp. 61–102. ISBN 978-0-470-97583-1. [Google Scholar]
- Puentes-Téllez, P.E.; Falcao Salles, J. Construction of Effective Minimal Active Microbial Consortia for Lignocellulose Degradation. Microb. Ecol. 2018, 76, 419–429. [Google Scholar] [CrossRef]
- Zhao, X.; Zhang, L.; Liu, D. Biomass recalcitrance. Part I: The chemical compositions and physical structures affecting the enzymatic hydrolysis of lignocellulose. Biofuels Bioprod. Biorefining 2012, 6, 465–482. [Google Scholar] [CrossRef]
- Bayer, E.A.; Shoham, Y.; Lamed, R. Lignocellulose-Decomposing Bacteria and Their Enzyme Systems. In The Prokaryotes; Rosenberg, E., DeLong, E.F., Lory, S., Stackebrandt, E., Thompson, F., Eds.; Springer: Berlin/Heidelberg, Germany, 2013; pp. 215–266. ISBN 978-3-642-30140-7. [Google Scholar]
- French, A.D. Glucose, not cellobiose, is the repeating unit of cellulose and why that is important. Cellulose 2017, 24, 4605–4609. [Google Scholar] [CrossRef]
- Zhou, X.; Li, W.; Mabon, R.; Broadbelt, L.J. A Critical Review on Hemicellulose Pyrolysis. Energy Technol. 2017, 5, 52–79. [Google Scholar] [CrossRef]
- Ridley, B.L.; O’Neill, M.A.; Mohnen, D. Pectins: Structure, biosynthesis, and oligogalacturonide-related signaling. Phytochemistry 2001, 57, 929–967. [Google Scholar] [CrossRef]
- dos Santos, A.C.; Ximenes, E.; Kim, Y.; Ladisch, M.R. Lignin–Enzyme Interactions in the Hydrolysis of Lignocellulosic Biomass. Trends Biotechnol. 2019, 37, 518–531. [Google Scholar] [CrossRef]
- De Carvalho, D.M.; Sevastyanova, O.; Penna, L.S.; da Silva, B.P.; Lindström, M.E.; Colodette, J.L. Assessment of chemical transformations in eucalyptus, sugarcane bagasse and straw during hydrothermal, dilute acid, and alkaline pretreatments. Ind. Crop. Prod. 2015, 73, 118–126. [Google Scholar] [CrossRef]
- Kamimura, N.; Sakamoto, S.; Mitsuda, N.; Masai, E.; Kajita, S. Advances in microbial lignin degradation and its applications. Curr. Opin. Biotechnol. 2019, 56, 179–186. [Google Scholar] [CrossRef] [PubMed]
- Wilhelm, R.C.; Singh, R.; Eltis, L.D.; Mohn, W.W. Bacterial contributions to delignification and lignocellulose degradation in forest soils with metagenomic and quantitative stable isotope probing. ISME J. 2019, 13, 413–429. [Google Scholar] [CrossRef]
- De Boer, W.; Folman, L.B.; Summerbell, R.C.; Boddy, L. Living in a fungal world: Impact of fungi on soil bacterial niche development. FEMS Microbiol. Rev. 2005, 29, 795–811. [Google Scholar] [CrossRef]
- Sweeney, M.D.; Xu, F. Biomass Converting Enzymes as Industrial Biocatalysts for Fuels and Chemicals: Recent Developments. Catalysts 2012, 2, 244–263. [Google Scholar] [CrossRef]
- Beckham, G.T.; Johnson, C.W.; Karp, E.M.; Salvachúa, D.; Vardon, D.R. Opportunities and challenges in biological lignin valorization. Curr. Opin. Biotechnol. 2016, 42, 40–53. [Google Scholar] [CrossRef]
- Lynd, L.R.; Weimer, P.J.; Van Zyl, W.H.; Pretorius, I.S. Microbial Cellulose Utilization: Fundamentals and Biotechnology. Microbiol. Mol. Biol. Rev. MMBT 2002, 66, 506–577. [Google Scholar] [CrossRef] [PubMed]
- Constancio, M.T.L.; Sacco, L.P.; Campanharo, J.C.; Castellane, T.C.L.; de Oliveira Souza, A.C.; Weiss, B.; de Mello Varani, A.; Alves, L.M.C. Exploring the Potential of Two Bacterial Consortia to Degrade Cellulosic Biomass for Biotechnological Applications. Curr. Microbiol. 2020, 77, 3114–3124. [Google Scholar] [CrossRef]
- De Jesus, R.B.; Omori, W.P.; de Macedo Lemos, E.G.; de Souza, J.A.M. Bacterial diversity in bovine rumen by metagenomic 16S rDNA sequencing and scanning electron microscopy. Acta Sci. Anim. Sci. 2015, 37, 251–257. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Goering, H.K. Forage Fiber Analyses: (Apparatus, Reagents, Procedures, and Some Applications); Agricultural Research Service, U.S. Dept. of Agriculture: Washington, DC, USA, 1970.
- Zhu, Y.; Malten, M.; Torry-Smith, M.; McMillan, J.D.; Stickel, J.J. Calculating sugar yields in high solids hydrolysis of biomass. Bioresour. Technol. 2011, 102, 2897–2903. [Google Scholar] [CrossRef] [PubMed]
- Uritskiy, G.V.; DiRuggiero, J.; Taylor, J. MetaWRAP—A flexible pipeline for genome-resolved metagenomic data analysis. Microbiome 2018, 6, 158. [Google Scholar] [CrossRef]
- Li, D.; Liu, C.-M.; Luo, R.; Sadakane, K.; Lam, T.-W. MEGAHIT: An ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 2015, 31, 1674–1676. [Google Scholar] [CrossRef]
- Parks, D.H.; Imelfort, M.; Skennerton, C.T.; Hugenholtz, P.; Tyson, G.W. CheckM: Assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015, 25, 1043–1055. [Google Scholar] [CrossRef]
- Chaumeil, P.-A.; Mussig, A.J.; Hugenholtz, P.; Parks, D.H. GTDB-Tk: A toolkit to classify genomes with the Genome Taxonomy Database. Bioinformatics 2019, 36, 1925–1927. [Google Scholar] [CrossRef]
- Wood, D.E.; Salzberg, S.L. Kraken: Ultrafast metagenomic sequence classification using exact alignments. Genome Biol. 2014, 15, R46. [Google Scholar] [CrossRef]
- O’Leary, N.A.; Wright, M.W.; Brister, J.R.; Ciufo, S.; Haddad, D.; McVeigh, R.; Rajput, B.; Robbertse, B.; Smith-White, B.; Ako-Adjei, D.; et al. Reference sequence (RefSeq) database at NCBI: Current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016, 44, D733–D745. [Google Scholar] [CrossRef] [PubMed]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST Server: Rapid Annotations using Subsystems Technology. BMC Genom. 2008, 9, 75. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Sato, Y.; Morishima, K. BlastKOALA and GhostKOALA: KEGG Tools for Functional Characterization of Genome and Metagenome Sequences. J. Mol. Biol. 2016, 428, 726–731. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Yohe, T.; Huang, L.; Entwistle, S.; Wu, P.; Yang, Z.; Busk, P.K.; Xu, Y.; Yin, Y. dbCAN2: A meta server for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 2018, 46, W95–W101. [Google Scholar] [CrossRef]
- Huerta-Cepas, J.; Szklarczyk, D.; Heller, D.; Hernández-Plaza, A.; Forslund, S.K.; Cook, H.; Mende, D.R.; Letunic, I.; Rattei, T.; Jensen, L.J.; et al. eggNOG 5.0: A hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res. 2019, 47, D309–D314. [Google Scholar] [CrossRef]
- McDaniel, E.A.; Anantharaman, K.; McMahon, K.D. MetabolisHMM: Phylogenomic Analysis for Exploration of Microbial Phylogenies and Metabolic Pathways. bioRxiv 2020. [Google Scholar] [CrossRef]
- Page, A.J.; Cummins, C.A.; Hunt, M.; Wong, V.K.; Reuter, S.; Holden, M.T.G.; Fookes, M.; Falush, D.; Keane, J.A.; Parkhill, J. Roary: Rapid large-scale prokaryote pan genome analysis. Bioinformatics 2015, 31, 3691–3693. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef] [PubMed]
- Gupta, V.K.; Tuohy, M.G. (Eds.) Biofuel Technologies: Recent Developments; Springer: Berlin/Heidelberg, Germany, 2013; ISBN 978-3-642-34518-0. [Google Scholar]
- Jiménez, D.J.; Dini-Andreote, F.; DeAngelis, K.M.; Singer, S.W.; Salles, J.F.; Van Elsas, J.D. Ecological Insights into the Dynamics of Plant Biomass-Degrading Microbial Consortia. Trends Microbiol. 2017, 25, 788–796. [Google Scholar] [CrossRef]
- Carlos, C.; Fan, H.; Currie, C.R. Substrate Shift Reveals Roles for Members of Bacterial Consortia in Degradation of Plant Cell Wall Polymers. Front. Microbiol. 2018, 9, 364. [Google Scholar] [CrossRef]
- Xu, R.; Zhang, K.; Liu, P.; Han, H.; Zhao, S.; Kakade, A.; Khan, A.; Du, D.; Li, X. Lignin depolymerization and utilization by bacteria. Bioresour. Technol. 2018, 269, 557–566. [Google Scholar] [CrossRef] [PubMed]
- Ventorino, V.; Aliberti, A.; Faraco, V.; Robertiello, A.; Giacobbe, S.; Ercolini, D.; Amore, A.; Fagnano, M.; Pepe, O. Exploring the microbiota dynamics related to vegetable biomasses degradation and study of lignocellulose-degrading bacteria for industrial biotechnological application. Sci. Rep. 2015, 5, 8161. [Google Scholar] [CrossRef]
- Puentes-Téllez, P.E.; Salles, J.F. Dynamics of Abundant and Rare Bacteria during Degradation of Lignocellulose from Sugarcane Biomass. Microb. Ecol. 2020, 79, 312–325. [Google Scholar] [CrossRef]
- Hu, F.; Jung, S.; Ragauskas, A. Pseudo-lignin formation and its impact on enzymatic hydrolysis. Bioresour. Technol. 2012, 117, 7–12. [Google Scholar] [CrossRef] [PubMed]
- Brenner, K.; You, L.; Arnold, F.H. Engineering microbial consortia: A new frontier in synthetic biology. Trends Biotechnol. 2008, 26, 483–489. [Google Scholar] [CrossRef]
- Tzamali, E.; Poirazi, P.; Tollis, I.G.; Reczko, M. A computational exploration of bacterial metabolic diversity identifying metabolic interactions and growth-efficient strain communities. BMC Syst. Biol. 2011, 5, 167. [Google Scholar] [CrossRef] [PubMed]
- Henrissat, B. A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem. J. 1991, 280, 309–316. [Google Scholar] [CrossRef]
- Sinnott, M.L. Catalytic mechanism of enzymic glycosyl transfer. Chem. Rev. 1990, 90, 1171–1202. [Google Scholar] [CrossRef]
- Boraston, A.B.; Healey, M.; Klassen, J.; Ficko-Blean, E.; van Bueren, A.L.; Law, V. A Structural and Functional Analysis of α-Glucan Recognition by Family 25 and 26 Carbohydrate-binding Modules Reveals a Conserved Mode of Starch Recognition. J. Biol. Chem. 2006, 281, 587–598. [Google Scholar] [CrossRef]
- Gilkes, N.R.; Warren, R.A.; Miller, R.C.; Kilburn, D.G. Precise Excision of the Cellulose Binding Domains from Two Cellulomonas Fimi Cellulases by a Homologous Protease and the Effect on Catalysis. J. Biol. Chem. 1988, 263, 10401–10407. [Google Scholar] [CrossRef]
- Black, G.W.; Rixon, J.E.; Clarke, J.H.; Hazlewood, G.P.; Theodorou, M.K.; Morris, P.; Gilbert, H.J. Evidence that linker sequences and cellulose-binding domains enhance the activity of hemicellulases against complex substrates. Biochem. J. 1996, 319, 515–520. [Google Scholar] [CrossRef]
- Cantarel, B.L.; Coutinho, P.M.; Rancurel, C.; Bernard, T.; Lombard, V.; Henrissat, B. The Carbohydrate-Active EnZymes database (CAZy): An expert resource for Glycogenomics. Nucleic Acids Res. 2009, 37, D233–D238. [Google Scholar] [CrossRef]
- Christov, L.P.; Prior, B.A. Esterases of xylan-degrading microorganisms: Production, properties, and significance. Enzym. Microb. Technol. 1993, 15, 460–475. [Google Scholar] [CrossRef]
- Michaud, P.; Da Costa, A.; Courtois, B.; Courtois, J. Polysaccharide Lyases: Recent Developments as Biotechnological Tools. Crit. Rev. Biotechnol. 2003, 23, 233–266. [Google Scholar] [CrossRef]
- Levasseur, A.; Drula, E.; Lombard, V.; Coutinho, P.M.; Henrissat, B. Expansion of the enzymatic repertoire of the CAZy database to integrate auxiliary redox enzymes. Biotechnol. Biofuels 2013, 6, 41. [Google Scholar] [CrossRef]
- Perez-Garcia, O.; Lear, G.; Singhal, N. Metabolic Network Modeling of Microbial Interactions in Natural and Engineered Environmental Systems. Front. Microbiol. 2016, 7, 673. [Google Scholar] [CrossRef] [PubMed]
- Fang, X.; Lloyd, C.J.; Palsson, B.O. Reconstructing organisms in silico: Genome-scale models and their emerging applications. Nat. Rev. Microbiol. 2020, 1–13. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Weiss, B.; Souza, A.C.O.; Constancio, M.T.L.; Alvarenga, D.O.; Pylro, V.S.; Alves, L.M.C.; Varani, A.M. Unraveling a Lignocellulose-Decomposing Bacterial Consortium from Soil Associated with Dry Sugarcane Straw by Genomic-Centered Metagenomics. Microorganisms 2021, 9, 995. https://doi.org/10.3390/microorganisms9050995
Weiss B, Souza ACO, Constancio MTL, Alvarenga DO, Pylro VS, Alves LMC, Varani AM. Unraveling a Lignocellulose-Decomposing Bacterial Consortium from Soil Associated with Dry Sugarcane Straw by Genomic-Centered Metagenomics. Microorganisms. 2021; 9(5):995. https://doi.org/10.3390/microorganisms9050995
Chicago/Turabian StyleWeiss, Bruno, Anna Carolina Oliveira Souza, Milena Tavares Lima Constancio, Danillo Oliveira Alvarenga, Victor S. Pylro, Lucia M. Carareto Alves, and Alessandro M. Varani. 2021. "Unraveling a Lignocellulose-Decomposing Bacterial Consortium from Soil Associated with Dry Sugarcane Straw by Genomic-Centered Metagenomics" Microorganisms 9, no. 5: 995. https://doi.org/10.3390/microorganisms9050995
APA StyleWeiss, B., Souza, A. C. O., Constancio, M. T. L., Alvarenga, D. O., Pylro, V. S., Alves, L. M. C., & Varani, A. M. (2021). Unraveling a Lignocellulose-Decomposing Bacterial Consortium from Soil Associated with Dry Sugarcane Straw by Genomic-Centered Metagenomics. Microorganisms, 9(5), 995. https://doi.org/10.3390/microorganisms9050995






