Biological Functions of Type II Toxin-Antitoxin Systems in Bacteria
Abstract
:1. Introduction
2. TA Systems Biology and Classification
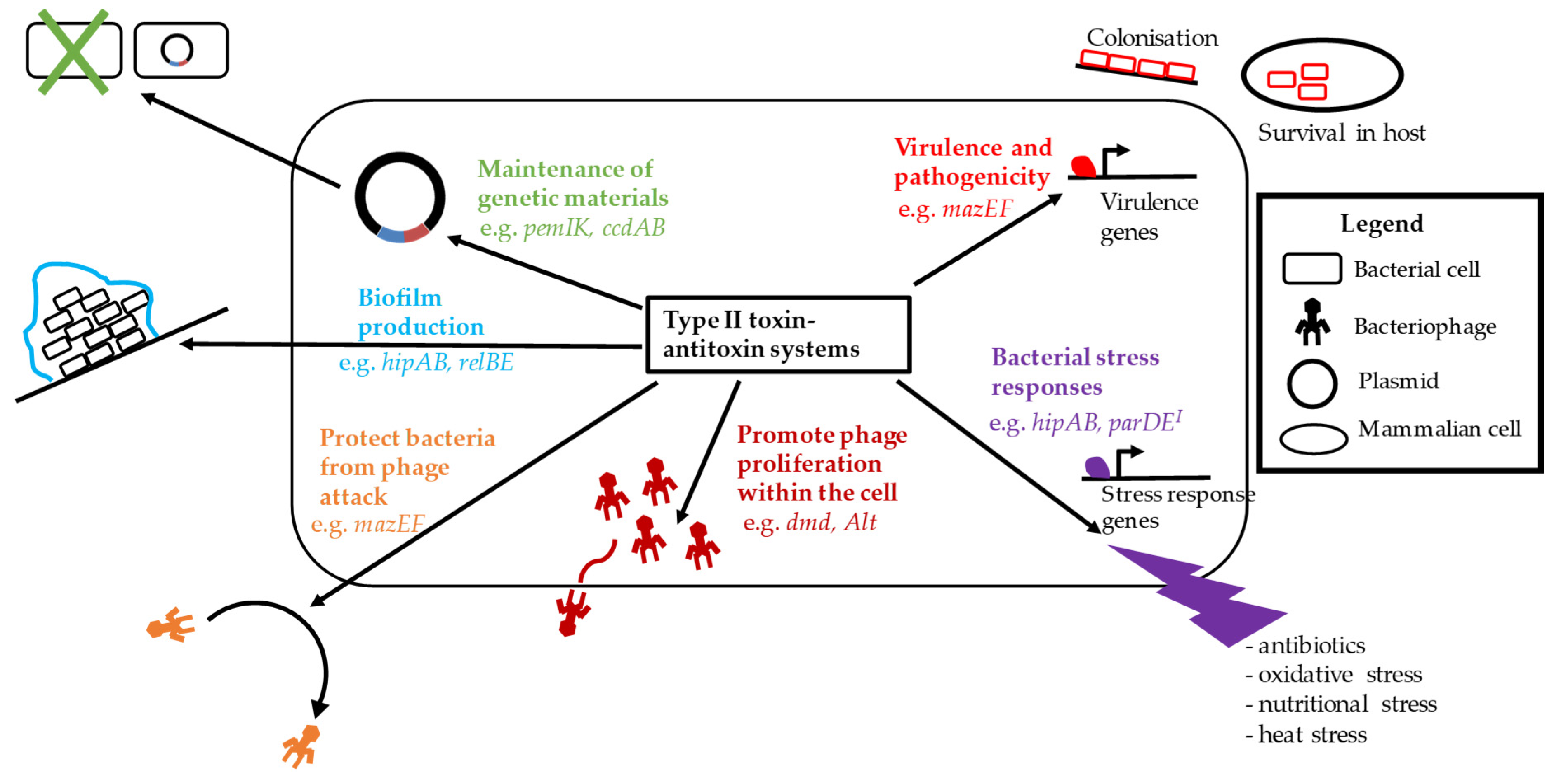
3. Biological Functions of Type II TA Systems
3.1. Maintenance of Genetic Materials
3.1.1. Plasmid Maintenance
3.1.2. Maintenance of Other Genetic Elements
3.2. Type II TAS in Bacterial Virulence and Pathogenesis
3.2.1. Virulence and Pathogenesis Mediated by Plasmid-Borne Type II TA Systems
3.2.2. Virulence and Pathogenesis Mediated by Chromosomal Type II TA Systems
3.2.3. Type II TA Systems That Negatively Regulate Virulence
3.3. Type II TAS Associated with Bacterial Biofilm Formation
3.4. Role of Type II TA Systems in Bacteriophage Resistance to Bacteria
3.5. Bacteriophage-Borne Type II TA or Antitoxin Helps Phage Propagation in the Host Bacteria
3.6. Stress Responses Mediated by Type II TA Systems
3.6.1. Antibiotic Tolerance and Persister Formation
3.6.2. Oxidative Stress
3.6.3. Nutritional Stress
3.6.4. Heat Tolerance
3.6.5. Other Stresses
4. Applications of Type II TA Systems in Biotechnology and Medicine
5. Limitations in TA Research
6. Concluding Remarks
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hayes, F. Toxins-antitoxins: Plasmid maintenance, programmed cell death, and cell cycle arrest. Science 2003, 301, 1496–1499. [Google Scholar] [CrossRef]
- Unterholzner, S.J.; Poppenberger, B.; Rozhon, W. Toxin-antitoxin systems: Biology, identification, and application. Mob. Genet. Elem. 2013, 3, e26219. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Melderen, L. Toxin-antitoxin systems: Why so many, what for? Curr. Opin. Microbiol. 2010, 13, 781–785. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, Y.; Park, J.H.; Inouye, M. Toxin-antitoxin systems in bacteria and archaea. Annu. Rev. Genet. 2011, 45, 61–79. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ogura, T.; Hiraga, S. Mini-F plasmid genes that couple host cell division to plasmid proliferation. Proc. Natl. Acad. Sci. USA 1983, 80, 4784–4788. [Google Scholar] [CrossRef] [Green Version]
- Akarsu, H.; Bordes, P.; Mansour, M.; Bigot, D.J.; Genevaux, P.; Falquet, L. TASmania: A bacterial Toxin-Antitoxin Systems database. PLoS Comput. Biol. 2019, 15, e1006946. [Google Scholar] [CrossRef] [Green Version]
- Shao, Y.; Harrison, E.M.; Bi, D.; Tai, C.; He, X.; Ou, H.Y.; Rajakumar, K.; Deng, Z. TADB: A web-based resource for Type 2 toxin-antitoxin loci in bacteria and archaea. Nucleic Acids Res. 2011, 39, D606–D611. [Google Scholar] [CrossRef] [Green Version]
- Kamruzzaman, M.; Iredell, J. A ParDE-family toxin antitoxin system in major resistance plasmids of Enterobacteriaceae confers antibiotic and heat tolerance. Sci. Rep. 2019, 9, 9872. [Google Scholar] [CrossRef] [Green Version]
- Lobato-Marquez, D.; Diaz-Orejas, R.; Garcia-Del Portillo, F. Toxin-antitoxins and bacterial virulence. FEMS Microbiol. Rev. 2016, 40, 592–609. [Google Scholar] [CrossRef] [Green Version]
- Page, R.; Peti, W. Toxin-antitoxin systems in bacterial growth arrest and persistence. Nat. Chem. Biol. 2016, 12, 208–214. [Google Scholar] [CrossRef]
- Tripathi, A.; Dewan, P.C.; Barua, B.; Varadarajan, R. Additional role for the ccd operon of F-plasmid as a transmissible persistence factor. Proc. Natl. Acad. Sci. USA 2012, 109, 12497–12502. [Google Scholar] [CrossRef] [Green Version]
- Wen, Y.; Behiels, E.; Devreese, B. Toxin-Antitoxin systems: Their role in persistence, biofilm formation, and pathogenicity. Pathog. Dis. 2014, 70, 240–249. [Google Scholar] [CrossRef]
- Yamaguchi, Y.; Inouye, M. Regulation of growth and death in Escherichia coli by toxin-antitoxin systems. Nat. Rev. Microbiol. 2011, 9, 779–790. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.E.; Walsh, T.R. Toxin-antitoxin systems and their role in disseminating and maintaining antimicrobial resistance. FEMS Microbiol. Rev. 2017, 41, 343–353. [Google Scholar] [CrossRef]
- Zhang, S.-P.; Wang, Q.; Quan, S.-W.; Yu, X.-Q.; Wang, Y.; Guo, D.-D.; Peng, L.; Feng, H.-Y.; He, Y.-X. Type II toxin–antitoxin system in bacteria: Activation, function, and mode of action. Biophys. Rep. 2020, 6, 68–79. [Google Scholar] [CrossRef]
- Marimon, O.; Teixeira, J.M.; Cordeiro, T.N.; Soo, V.W.; Wood, T.L.; Mayzel, M.; Amata, I.; Garcia, J.; Morera, A.; Gay, M.; et al. An oxygen-sensitive toxin-antitoxin system. Nat. Commun. 2016, 7, 13634. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Lord, D.M.; Cheng, H.Y.; Osbourne, D.O.; Hong, S.H.; Sanchez-Torres, V.; Quiroga, C.; Zheng, K.; Herrmann, T.; Peti, W.; et al. A new type V toxin-antitoxin system where mRNA for toxin GhoT is cleaved by antitoxin GhoS. Nat. Chem. Biol. 2012, 8, 855–861. [Google Scholar] [CrossRef] [Green Version]
- Aakre, C.D.; Phung, T.N.; Huang, D.; Laub, M.T. A bacterial toxin inhibits DNA replication elongation through a direct interaction with the beta sliding clamp. Mol. Cell 2013, 52, 617–628. [Google Scholar] [CrossRef] [Green Version]
- Gerdes, K.; Bech, F.W.; Jorgensen, S.T.; Lobner-Olesen, A.; Rasmussen, P.B.; Atlung, T.; Boe, L.; Karlstrom, O.; Molin, S.; von Meyenburg, K. Mechanism of postsegregational killing by the hok gene product of the parB system of plasmid R1 and its homology with the relF gene product of the E. coli relB operon. EMBO J. 1986, 5, 2023–2029. [Google Scholar] [CrossRef]
- Jaffe, A.; Ogura, T.; Hiraga, S. Effects of the ccd function of the F plasmid on bacterial growth. J. Bacteriol. 1985, 163, 841–849. [Google Scholar] [CrossRef] [Green Version]
- Gerdes, K.; Rasmussen, P.B.; Molin, S. Unique type of plasmid maintenance function: Postsegregational killing of plasmid-free cells. Proc. Natl. Acad. Sci. USA 1986, 83, 3116–3120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Couturier, M.; Bahassi el, M.; Van Melderen, L. Bacterial death by DNA gyrase poisoning. Trends Microbiol. 1998, 6, 269–275. [Google Scholar] [CrossRef]
- Duprilot, M.; Decre, D.; Genel, N.; Drieux, L.; Sougakoff, W.; Arlet, G. Diversity and functionality of plasmid-borne vagCD toxin-antitoxin systems of Klebsiella pneumoniae. J. Antimicrob. Chemother. 2017, 72, 1320–1326. [Google Scholar] [CrossRef]
- Engelberg-Kulka, H.; Glaser, G. Addiction modules and programmed cell death and antideath in bacterial cultures. Annu. Rev. Microbiol. 1999, 53, 43–70. [Google Scholar] [CrossRef] [PubMed]
- Gotfredsen, M.; Gerdes, K. The Escherichia coli relBE genes belong to a new toxin-antitoxin gene family. Mol. Microbiol. 1998, 29, 1065–1076. [Google Scholar] [CrossRef]
- Jensen, R.B.; Gerdes, K. Programmed cell death in bacteria: Proteic plasmid stabilization systems. Mol. Microbiol. 1995, 17, 205–210. [Google Scholar] [CrossRef]
- Li, Z.; Song, Q.; Wang, Y.; Xiao, X.; Xu, J. Identification of a functional toxin-antitoxin system located in the genomic island PYG1 of piezophilic hyperthermophilic archaeon Pyrococcus yayanosii. Extremophiles 2018, 22, 347–357. [Google Scholar] [CrossRef]
- Chan, W.T.; Espinosa, M.; Yeo, C.C. Keeping the Wolves at Bay: Antitoxins of prokaryotic type II toxin-antitoxin systems. Front. Mol. Biosci. 2016, 3, 9. [Google Scholar] [CrossRef] [Green Version]
- Cherny, I.; Rockah, L.; Gazit, E. The YoeB toxin is a folded protein that forms a physical complex with the unfolded YefM antitoxin. Implications for a structural-based differential stability of toxin-antitoxin systems. J. Biol. Chem. 2005, 280, 30063–30072. [Google Scholar] [CrossRef] [Green Version]
- Muthuramalingam, M.; White, J.C.; Bourne, C.R. Toxin-antitoxin modules are pliable switches activated by multiple protease pathways. Toxins 2016, 8, 214. [Google Scholar] [CrossRef]
- Song, S.; Wood, T.K. Toxin/antitoxin system paradigms: Toxins bound to antitoxins are not likely activated by preferential antitoxin degradation. Adv. Biosyst. 2020, 4, e1900290. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Lord, D.M.; Hong, S.H.; Peti, W.; Benedik, M.J.; Page, R.; Wood, T.K. Type II toxin/antitoxin MqsR/MqsA controls type V toxin/antitoxin GhoT/GhoS. Environ. Microbiol. 2013, 15, 1734–1744. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, A.Y.; Kamruzzaman, M.; Iredell, J.R. Specialised functions of two common plasmid mediated toxin-antitoxin systems, ccdAB and pemIK, in Enterobacteriaceae. PLoS ONE 2020, 15, e0230652. [Google Scholar] [CrossRef] [PubMed]
- Di Cesare, A.; Losasso, C.; Barco, L.; Eckert, E.M.; Conficoni, D.; Sarasini, G.; Corno, G.; Ricci, A. Diverse distribution of Toxin-Antitoxin II systems in Salmonella enterica serovars. Sci. Rep. 2016, 6, 28759. [Google Scholar] [CrossRef] [Green Version]
- Engelberg-Kulka, H.; Hazan, R.; Amitai, S. mazEF: A chromosomal toxin-antitoxin module that triggers programmed cell death in bacteria. J. Cell Sci. 2005, 118, 4327–4332. [Google Scholar] [CrossRef] [Green Version]
- Gupta, K.; Tripathi, A.; Sahu, A.; Varadarajan, R. Contribution of the chromosomal ccdAB operon to bacterial drug tolerance. J. Bacteriol. 2017, 199. [Google Scholar] [CrossRef] [Green Version]
- Juhas, M.; van der Meer, J.R.; Gaillard, M.; Harding, R.M.; Hood, D.W.; Crook, D.W. Genomic islands: Tools of bacterial horizontal gene transfer and evolution. FEMS Microbiol. Rev. 2009, 33, 376–393. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yao, X.; Chen, T.; Shen, X.; Zhao, Y.; Wang, M.; Rao, X.; Yin, S.; Wang, J.; Gong, Y.; Lu, S.; et al. The chromosomal SezAT toxin-antitoxin system promotes the maintenance of the SsPI-1 pathogenicity island in epidemic Streptococcus suis. Mol. Microbiol. 2015, 98, 243–257. [Google Scholar] [CrossRef]
- Huguet, K.T.; Gonnet, M.; Doublet, B.; Cloeckaert, A. A toxin antitoxin system promotes the maintenance of the IncA/C-mobilizable Salmonella Genomic Island 1. Sci. Rep. 2016, 6, 32285. [Google Scholar] [CrossRef] [Green Version]
- Wozniak, R.A.; Waldor, M.K. A toxin-antitoxin system promotes the maintenance of an integrative conjugative element. PLoS Genet 2009, 5, e1000439. [Google Scholar] [CrossRef]
- Yuan, J.; Yamaichi, Y.; Waldor, M.K. The three vibrio cholerae chromosome II-encoded ParE toxins degrade chromosome I following loss of chromosome II. J. Bacteriol. 2011, 193, 611–619. [Google Scholar] [CrossRef] [Green Version]
- Szekeres, S.; Dauti, M.; Wilde, C.; Mazel, D.; Rowe-Magnus, D.A. Chromosomal toxin-antitoxin loci can diminish large-scale genome reductions in the absence of selection. Mol. Microbiol. 2007, 63, 1588–1605. [Google Scholar] [CrossRef]
- Klein, S.; Pipes, S.; Lovell, C.R. Occurrence and significance of pathogenicity and fitness islands in environmental vibrios. AMB Express 2018, 8, 177. [Google Scholar] [CrossRef]
- Bustamante, P.; Vidal, R. Repertoire and diversity of toxin–antitoxin systems of crohn’s disease-associated adherent-invasive Escherichia coli. New insight of this emergent E. coli pathotype. Front. Microbiol. 2020, 11, 807. [Google Scholar] [CrossRef] [PubMed]
- Lima-Mendez, G.; Oliveira Alvarenga, D.; Ross, K.; Hallet, B.; Van Melderen, L.; Varani, A.M.; Chandler, M. Toxin-antitoxin gene pairs found in Tn3 family transposons appear to be an integral part of the transposition module. mBio 2020, 11, e00452-20. [Google Scholar] [CrossRef] [Green Version]
- Bustamante, P.; Tello, M.; Orellana, O. Toxin-antitoxin systems in the mobile genome of Acidithiobacillus ferrooxidans. PLoS ONE 2014, 9, e112226. [Google Scholar] [CrossRef] [Green Version]
- Yao, J.; Guo, Y.; Wang, P.; Zeng, Z.; Li, B.; Tang, K.; Liu, X.; Wang, X. Type II toxin/antitoxin system ParESO /CopASO stabilizes prophage CP4So in Shewanella oneidensis. Environ. Microbiol. 2018, 20, 1224–1239. [Google Scholar] [CrossRef] [PubMed]
- Ramage, H.R.; Connolly, L.E.; Cox, J.S. Comprehensive functional analysis of Mycobacterium tuberculosis toxin-antitoxin systems: Implications for pathogenesis, stress responses, and evolution. PLoS Genet. 2009, 5, e1000767. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De la Cruz, M.A.; Zhao, W.; Farenc, C.; Gimenez, G.; Raoult, D.; Cambillau, C.; Gorvel, J.P.; Meresse, S. A toxin-antitoxin module of Salmonella promotes virulence in mice. PLoS Pathog. 2013, 9, e1003827. [Google Scholar] [CrossRef]
- McVicker, G.; Tang, C.M. Deletion of toxin-antitoxin systems in the evolution of Shigella sonnei as a host-adapted pathogen. Nat. Microbiol. 2016, 2, 16204. [Google Scholar] [CrossRef] [PubMed]
- Sayeed, S.; Brendler, T.; Davis, M.; Reaves, L.; Austin, S. Surprising dependence on postsegregational killing of host cells for maintenance of the large virulence plasmid of Shigella flexneri. J. Bacteriol. 2005, 187, 2768–2773. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lobato-Marquez, D.; Molina-Garcia, L.; Moreno-Cordoba, I.; Garcia-Del Portillo, F.; Diaz-Orejas, R. Stabilization of the virulence plasmid pSLT of Salmonella Typhimurium by three maintenance systems and its evaluation by using a new stability test. Front. Mol. Biosci. 2016, 3, 66. [Google Scholar] [CrossRef] [Green Version]
- Lobato-Marquez, D.; Moreno-Cordoba, I.; Figueroa, V.; Diaz-Orejas, R.; Garcia-del Portillo, F. Distinct type I and type II toxin-antitoxin modules control Salmonella lifestyle inside eukaryotic cells. Sci. Rep. 2015, 5, 9374. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hernandez-Ramirez, K.C.; Chavez-Jacobo, V.M.; Valle-Maldonado, M.I.; Patino-Medina, J.A.; Diaz-Perez, S.P.; Jacome-Galarza, I.E.; Ortiz-Alvarado, R.; Meza-Carmen, V.; Ramirez-Diaz, M.I. Plasmid pUM505 encodes a Toxin-Antitoxin system conferring plasmid stability and increased Pseudomonas aeruginosa virulence. Microb. Pathog. 2017, 112, 259–268. [Google Scholar] [CrossRef]
- Hernandez-Ramirez, K.C.; Valerio-Arellano, B.; Valle-Maldonado, M.I.; Ruiz-Herrera, L.F.; Meza-Carmen, V.; Ramirez-Diaz, M.I. Virulence conferred by PumA toxin from the plasmid-encoded PumAB toxin-antitoxin system is regulated by quorum system. Curr. Microbiol. 2020, 77, 2535–2543. [Google Scholar] [CrossRef]
- Bukowski, M.; Rojowska, A.; Wladyka, B. Prokaryotic toxin-antitoxin systems—The role in bacterial physiology and application in molecular biology. Acta Biochim. Pol. 2011, 58, 1–9. [Google Scholar] [CrossRef]
- Brown, J.S.; Gilliland, S.M.; Spratt, B.G.; Holden, D.W. A locus contained within a variable region of pneumococcal pathogenicity island 1 contributes to virulence in mice. Infect. Immun. 2004, 72, 1587–1593. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mutschler, H.; Meinhart, A. epsilon/zeta systems: Their role in resistance, virulence, and their potential for antibiotic development. J. Mol. Med. 2011, 89, 1183–1194. [Google Scholar] [CrossRef] [Green Version]
- Wood, T.L.; Wood, T.K. The HigB/HigA toxin/antitoxin system of Pseudomonas aeruginosa influences the virulence factors pyochelin, pyocyanin, and biofilm formation. Microbiologyopen 2016, 5, 499–511. [Google Scholar] [CrossRef]
- Agarwal, S.; Tiwari, P.; Deep, A.; Kidwai, S.; Gupta, S.; Thakur, K.G.; Singh, R. System-wide analysis unravels the differential regulation and in vivo essentiality of virulence-associated proteins B and C toxin-antitoxin systems of Mycobacterium tuberculosis. J. Infect. Dis. 2018, 217, 1809–1820. [Google Scholar] [CrossRef]
- Deep, A.; Tiwari, P.; Agarwal, S.; Kaundal, S.; Kidwai, S.; Singh, R.; Thakur, K.G. Structural, functional and biological insights into the role of Mycobacterium tuberculosis VapBC11 toxin-antitoxin system: Targeting a tRNase to tackle mycobacterial adaptation. Nucleic Acids Res. 2018, 46, 11639–11655. [Google Scholar] [CrossRef] [Green Version]
- Tiwari, P.; Arora, G.; Singh, M.; Kidwai, S.; Narayan, O.P.; Singh, R. MazF ribonucleases promote Mycobacterium tuberculosis drug tolerance and virulence in guinea pigs. Nat. Commun. 2015, 6, 6059. [Google Scholar] [CrossRef] [Green Version]
- Ma, D.; Mandell, J.B.; Donegan, N.P.; Cheung, A.L.; Ma, W.; Rothenberger, S.; Shanks, R.M.Q.; Richardson, A.R.; Urish, K.L. The toxin-antitoxin MazEF drives Staphylococcus aureus biofilm formation, antibiotic tolerance, and chronic infection. mBio 2019, 10. [Google Scholar] [CrossRef] [Green Version]
- Ren, D.; Walker, A.N.; Daines, D.A. Toxin-antitoxin loci vapBC-1 and vapXD contribute to survival and virulence in nontypeable Haemophilus influenzae. BMC Microbiol. 2012, 12, 263. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ren, D.; Kordis, A.A.; Sonenshine, D.E.; Daines, D.A. The ToxAvapA toxin-antitoxin locus contributes to the survival of nontypeable Haemophilus influenzae during infection. PLoS ONE 2014, 9, e91523. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Norton, J.P.; Mulvey, M.A. Toxin-antitoxin systems are important for niche-specific colonization and stress resistance of uropathogenic Escherichia coli. PLoS Pathog. 2012, 8, e1002954. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, H.; Hay, A.J.; Zhong, Z.; Zhu, J.; Kan, B. Functional RelBE-Family toxin-antitoxin pairs affect biofilm maturation and intestine colonization in Vibrio cholerae. PLoS ONE 2015, 10, e0135696. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hopper, S.; Wilbur, J.S.; Vasquez, B.L.; Larson, J.; Clary, S.; Mehr, I.J.; Seifert, H.S.; So, M. Isolation of Neisseria gonorrhoeae mutants that show enhanced trafficking across polarized T84 epithelial monolayers. Infect. Immun. 2000, 68, 896–905. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martner, A.; Skovbjerg, S.; Paton, J.C.; Wold, A.E. Streptococcus pneumoniae autolysis prevents phagocytosis and production of phagocyte-activating cytokines. Infect. Immun. 2009, 77, 3826–3837. [Google Scholar] [CrossRef] [Green Version]
- Wen, W.; Liu, B.; Xue, L.; Zhu, Z.; Niu, L.; Sun, B. Autoregulation and virulence control by the toxin-antitoxin system SavRS in Staphylococcus aureus. Infect. Immun. 2018, 86, e00032-18. [Google Scholar] [CrossRef] [Green Version]
- Guo, Y.; Sun, C.; Li, Y.; Tang, K.; Ni, S.; Wang, X. Antitoxin HigA inhibits virulence gene mvfR expression in Pseudomonas aeruginosa. Environ. Microbiol. 2019, 21, 2707–2723. [Google Scholar] [CrossRef]
- Chaudhuri, R.R.; Morgan, E.; Peters, S.E.; Pleasance, S.J.; Hudson, D.L.; Davies, H.M.; Wang, J.; van Diemen, P.M.; Buckley, A.M.; Bowen, A.J.; et al. Comprehensive assignment of roles for Salmonella Typhimurium genes in intestinal colonization of food-producing animals. PLoS Genet. 2013, 9, e1003456. [Google Scholar] [CrossRef] [Green Version]
- Kung, V.L.; Khare, S.; Stehlik, C.; Bacon, E.M.; Hughes, A.J.; Hauser, A.R. An rhs gene of Pseudomonas aeruginosa encodes a virulence protein that activates the inflammasome. Proc. Natl. Acad. Sci. USA 2012, 109, 1275–1280. [Google Scholar] [CrossRef] [Green Version]
- Mulder, D.T.; Cooper, C.A.; Coombes, B.K. Type VI secretion system-associated gene clusters contribute to pathogenesis of Salmonella enterica serovar Typhimurium. Infect. Immun. 2012, 80, 1996–2007. [Google Scholar] [CrossRef] [Green Version]
- Starsta, M.; Hammarlof, D.L.; Waneskog, M.; Schlegel, S.; Xu, F.; Heden Gynna, A.; Borg, M.; Herschend, S.; Koskiniemi, S. RHS-elements function as type II toxin-antitoxin modules that regulate intra-macrophage replication of Salmonella Typhimurium. PLoS Genet. 2020, 16, e1008607. [Google Scholar] [CrossRef]
- Pupo, G.M.; Lan, R.; Reeves, P.R. Multiple independent origins of Shigella clones of Escherichia coli and convergent evolution of many of their characteristics. Proc. Natl. Acad. Sci. USA 2000, 97, 10567–10572. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bernardini, M.L.; Mounier, J.; d’Hauteville, H.; Coquis-Rondon, M.; Sansonetti, P.J. Identification of icsA, a plasmid locus of Shigella flexneri that governs bacterial intra- and intercellular spread through interaction with F-actin. Proc. Natl. Acad. Sci. USA 1989, 86, 3867–3871. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brotcke Zumsteg, A.; Goosmann, C.; Brinkmann, V.; Morona, R.; Zychlinsky, A. IcsA is a Shigella flexneri adhesin regulated by the type III secretion system and required for pathogenesis. Cell Host Microbe 2014, 15, 435–445. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bardaji, L.; Anorga, M.; Echeverria, M.; Ramos, C.; Murillo, J. The toxic guardians—Multiple toxin-antitoxin systems provide stability, avoid deletions and maintain virulence genes of Pseudomonas syringae virulence plasmids. Mob. DNA 2019, 10, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Doumith, M.; Dhanji, H.; Ellington, M.J.; Hawkey, P.; Woodford, N. Characterization of plasmids encoding extended-spectrum β-lactamases and their addiction systems circulating among Escherichia coli clinical isolates in the UK. J. Antimicrob. Chemother. 2012, 67, 878–885. [Google Scholar] [CrossRef] [Green Version]
- Mnif, B.; Harhour, H.; Jdidi, J.; Mahjoubi, F.; Genel, N.; Arlet, G.; Hammami, A. Molecular epidemiology of extended-spectrum β-lactamase-producing Escherichia coli in Tunisia and characterization of their virulence factors and plasmid addiction systems. BMC Microbiol. 2013, 13, 147. [Google Scholar] [CrossRef] [Green Version]
- Mnif, B.; Vimont, S.; Boyd, A.; Bourit, E.; Picard, B.; Branger, C.; Denamur, E.; Arlet, G. Molecular characterization of addiction systems of plasmids encoding extended-spectrum β-lactamases in Escherichia coli. J. Antimicrob. Chemother. 2010, 65, 1599–1603. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.; Venkataraman, B.; Vasudevan, M.; Gopinath Bankar, K. Co-expression network analysis of toxin-antitoxin loci in Mycobacterium tuberculosis reveals key modulators of cellular stress. Sci. Rep. 2017, 7, 5868. [Google Scholar] [CrossRef]
- Helaine, S.; Cheverton, A.M.; Watson, K.G.; Faure, L.M.; Matthews, S.A.; Holden, D.W. Internalization of Salmonella by macrophages induces formation of nonreplicating persisters. Science 2014, 343, 204–208. [Google Scholar] [CrossRef]
- Chimal-Cazares, F.; Hernandez-Martinez, G.; Pacheco, S.; Ares, M.A.; Soria-Bustos, J.; Sanchez-Gutierrez, M.; Izquierdo-Vega, J.A.; Ibarra, J.A.; Gonzalez, Y.M.J.A.; Gorvel, J.P.; et al. Molecular characterization of SehB, a type II antitoxin of Salmonella enterica serotype Typhimurium: Amino acid residues involved in DNA-binding, homodimerization, toxin interaction, and virulence. Front. Microbiol. 2020, 11, 614. [Google Scholar] [CrossRef] [PubMed]
- Molinaro, A.L.; Kashipathy, M.M.; Lovell, S.; Battaile, K.P.; Coussens, N.P.; Shen, M.; Daines, D.A. Crystal structure of VapBC-1 from nontypeable Haemophilus influenzae and the effect of pin domain mutations on survival during infection. J. Bacteriol. 2019, 201. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tettelin, H.; Nelson, K.E.; Paulsen, I.T.; Eisen, J.A.; Read, T.D.; Peterson, S.; Heidelberg, J.; DeBoy, R.T.; Haft, D.H.; Dodson, R.J.; et al. Complete genome sequence of a virulent isolate of Streptococcus pneumoniae. Science 2001, 293, 498–506. [Google Scholar] [CrossRef] [Green Version]
- Nau, R.; Eiffert, H. Modulation of release of proinflammatory bacterial compounds by antibacterials: Potential impact on course of inflammation and outcome in sepsis and meningitis. Clin. Microbiol. Rev. 2002, 15, 95–110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hirst, R.A.; Kadioglu, A.; O’Callaghan, C.; Andrew, P.W. The role of pneumolysin in pneumococcal pneumonia and meningitis. Clin. Exp. Immunol. 2004, 138, 195–201. [Google Scholar] [CrossRef] [PubMed]
- Cox, C.D. Effect of pyochelin on the virulence of Pseudomonas aeruginosa. Infect. Immun. 1982, 36, 17–23. [Google Scholar] [CrossRef] [Green Version]
- Sokol, P.A. Surface expression of ferripyochelin-binding protein is required for virulence of Pseudomonas aeruginosa. Infect. Immun. 1987, 55, 2021–2025. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Mushegian, A.; Lory, S.; Jin, S. Large-scale isolation of candidate virulence genes of Pseudomonas aeruginosa by in vivo selection. Proc. Natl. Acad. Sci. USA 1996, 93, 10434–10439. [Google Scholar] [CrossRef] [Green Version]
- Lau, G.W.; Hassett, D.J.; Ran, H.; Kong, F. The role of pyocyanin in Pseudomonas aeruginosa infection. Trends Mol. Med. 2004, 10, 599–606. [Google Scholar] [CrossRef]
- Kamruzzaman, M.; Udden, S.M.; Cameron, D.E.; Calderwood, S.B.; Nair, G.B.; Mekalanos, J.J.; Faruque, S.M. Quorum-regulated biofilms enhance the development of conditionally viable, environmental Vibrio cholerae. Proc. Natl. Acad. Sci. USA 2010, 107, 1588–1593. [Google Scholar] [CrossRef] [Green Version]
- Naser, I.B.; Hoque, M.M.; Faruque, S.N.; Kamruzzaman, M.; Yamasaki, S.; Faruque, S.M. Vibrio cholerae strains with inactivated cqsS gene overproduce autoinducer-2 which enhances resuscitation of dormant environmental V. cholerae. PLoS ONE 2019, 14, e0223226. [Google Scholar] [CrossRef] [PubMed]
- Bjarnsholt, T. The role of bacterial biofilms in chronic infections. APMIS 2013, 136, 1–51. [Google Scholar] [CrossRef]
- Mulcahy, L.R.; Isabella, V.M.; Lewis, K. Pseudomonas aeruginosa biofilms in disease. Microb. Ecol. 2014, 68, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Otto, M. Staphylococcal biofilms. Curr. Top. Microbiol. Immunol. 2008, 322, 207–228. [Google Scholar]
- Hall-Stoodley, L.; Stoodley, P. Developmental regulation of microbial biofilms. Curr. Opin. Biotechnol. 2002, 13, 228–233. [Google Scholar] [CrossRef]
- Gonzalez Barrios, A.F.; Zuo, R.; Hashimoto, Y.; Yang, L.; Bentley, W.E.; Wood, T.K. Autoinducer 2 controls biofilm formation in Escherichia coli through a novel motility quorum-sensing regulator (MqsR, B3022). J. Bacteriol. 2006, 188, 305–316. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, Y.; Wang, X.; Zhang, X.S.; Grigoriu, S.; Page, R.; Peti, W.; Wood, T.K. Escherichia coli toxin/antitoxin pair MqsR/MqsA regulate toxin CspD. Environ. Microbiol. 2010, 12, 1105–1121. [Google Scholar] [CrossRef] [Green Version]
- Ren, D.; Bedzyk, L.A.; Thomas, S.M.; Ye, R.W.; Wood, T.K. Gene expression in Escherichia coli biofilms. Appl. Microbiol. Biotechnol. 2004, 64, 515–524. [Google Scholar] [CrossRef] [PubMed]
- Soo, V.W.; Wood, T.K. Antitoxin MqsA represses curli formation through the master biofilm regulator CsgD. Sci. Rep. 2013, 3, 3186. [Google Scholar] [CrossRef] [Green Version]
- Fraikin, N.; Rousseau, C.J.; Goeders, N.; Van Melderen, L. Reassessing the role of the Type II MqsRA toxin-antitoxin system in stress response and biofilm formation: MqsA is transcriptionally uncoupled from mqsR. mBio 2019, 10. [Google Scholar] [CrossRef] [Green Version]
- Kolodkin-Gal, I.; Verdiger, R.; Shlosberg-Fedida, A.; Engelberg-Kulka, H. A differential effect of E. coli toxin-antitoxin systems on cell death in liquid media and biofilm formation. PLoS ONE 2009, 4, e6785. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, J.; Wang, Q.; Li, M.; Heijstra, B.D.; Wang, S.; Liang, Q.; Qi, Q. Escherichia coli toxin gene hipA affects biofilm formation and DNA release. Microbiology 2013, 159, 633–640. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bloom-Ackermann, Z.; Steinberg, N.; Rosenberg, G.; Oppenheimer-Shaanan, Y.; Pollack, D.; Ely, S.; Storzi, N.; Levy, A.; Kolodkin-Gal, I. Toxin-Antitoxin systems eliminate defective cells and preserve symmetry in Bacillus subtilis biofilms. Environ. Microbiol. 2016, 18, 5032–5047. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Zhao, X.; Wang, H.; Huang, X.; Duan, X.; Gu, Y.; Lambert, N.; Zhang, K.; Kou, Z.; Xie, J. Mycobacterium tuberculosis toxin Rv2872 is an RNase involved in vancomycin stress response and biofilm development. Appl. Microbiol. Biotechnol. 2018, 102, 7123–7133. [Google Scholar] [CrossRef]
- Zhang, Y.; Xia, B.; Li, M.; Shi, J.; Long, Y.; Jin, Y.; Bai, F.; Cheng, Z.; Jin, S.; Wu, W. HigB reciprocally controls biofilm formation and the expression of type III secretion system genes through influencing the intracellular c-di-GMP level in Pseudomonas aeruginosa. Toxins 2018, 10, 424. [Google Scholar] [CrossRef] [Green Version]
- Chan, W.T.; Domenech, M.; Moreno-Cordoba, I.; Navarro-Martinez, V.; Nieto, C.; Moscoso, M.; Garcia, E.; Espinosa, M. The Streptococcus pneumoniae yefM-yoeB and relBE toxin-antitoxin operons participate in oxidative stress and biofilm formation. Toxins 2018, 10, 378. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, Y.; Wang, X.; Ma, Q.; Zhang, X.S.; Wood, T.K. Toxin-antitoxin systems in Escherichia coli influence biofilm formation through YjgK (TabA) and fimbriae. J. Bacteriol. 2009, 191, 1258–1267. [Google Scholar] [CrossRef] [Green Version]
- Theunissen, S.; De Smet, L.; Dansercoer, A.; Motte, B.; Coenye, T.; Van Beeumen, J.J.; Devreese, B.; Savvides, S.N.; Vergauwen, B. The 285 kDa Bap/RTX hybrid cell surface protein (SO4317) of Shewanella oneidensis MR-1 is a key mediator of biofilm formation. Res. Microbiol. 2010, 161, 144–152. [Google Scholar] [CrossRef]
- Xu, J.; Xia, K.; Li, P.; Qian, C.; Li, Y.; Liang, X. Functional investigation of the chromosomal ccdAB and hipAB operon in Escherichia coli Nissle 1917. Appl. Microbiol. Biotechnol. 2020, 104, 6731–6747. [Google Scholar] [CrossRef] [PubMed]
- Salas-Jara, M.J.; Ilabaca, A.; Vega, M.; Garcia, A. Biofilm forming Lactobacillus: New challenges for the development of probiotics. Microorganisms 2016, 4, 35. [Google Scholar] [CrossRef] [PubMed]
- Ambalam, P.; Kondepudi, K.K.; Nilsson, I.; Wadstrom, T.; Ljungh, A. Bile enhances cell surface hydrophobicity and biofilm formation of bifidobacteria. Appl. Biochem. Biotechnol. 2014, 172, 1970–1981. [Google Scholar] [CrossRef] [PubMed]
- Van Acker, H.; Sass, A.; Dhondt, I.; Nelis, H.J.; Coenye, T. Involvement of toxin-antitoxin modules in Burkholderia cenocepacia biofilm persistence. Pathog. Dis. 2014, 71, 326–335. [Google Scholar] [CrossRef]
- Koga, M.; Otsuka, Y.; Lemire, S.; Yonesaki, T. Escherichia coli rnlA and rnlB compose a novel toxin-antitoxin system. Genetics 2011, 187, 123–130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Otsuka, Y.; Yonesaki, T. Dmd of bacteriophage T4 functions as an antitoxin against Escherichia coli LsoA and RnlA toxins. Mol. Microbiol. 2012, 83, 669–681. [Google Scholar] [CrossRef] [PubMed]
- Alawneh, A.M.; Qi, D.; Yonesaki, T.; Otsuka, Y. An ADP-ribosyltransferase Alt of bacteriophage T4 negatively regulates the Escherichia coli MazF toxin of a toxin-antitoxin module. Mol. Microbiol. 2016, 99, 188–198. [Google Scholar] [CrossRef]
- Kai, T.; Selick, H.E.; Yonesaki, T. Destabilization of bacteriophage T4 mRNAs by a mutation of gene 61.5. Genetics 1996, 144, 7–14. [Google Scholar] [CrossRef] [PubMed]
- Otsuka, Y.; Yonesaki, T. A novel endoribonuclease, RNase LS, in Escherichia coli. Genetics 2005, 169, 13–20. [Google Scholar] [CrossRef] [Green Version]
- Hosseini, N.; Pourhajibagher, M.; Chiniforush, N.; Hosseinkhan, N.; Rezaie, P.; Bahador, A. Modulation of toxin-antitoxin system RnlAB type II in phage-resistant Gammaproteobacteria surviving photodynamic treatment. J. Lasers Med. Sci. 2019, 10, 21–28. [Google Scholar] [CrossRef]
- Hazan, R.; Engelberg-Kulka, H. Escherichia coli mazEF-mediated cell death as a defense mechanism that inhibits the spread of phage P1. Mol. Genet. Genomics 2004, 272, 227–234. [Google Scholar] [CrossRef]
- Sberro, H.; Leavitt, A.; Kiro, R.; Koh, E.; Peleg, Y.; Qimron, U.; Sorek, R. Discovery of functional toxin/antitoxin systems in bacteria by shotgun cloning. Mol. Cell 2013, 50, 136–148. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, Y.; Gao, Z.; Zhang, H.; Dong, Y. Structural characterizations of phage antitoxin Dmd and its interactions with bacterial toxin RnlA. Biochem. Biophys. Res. Commun. 2016, 472, 592–597. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Liu, X.; Tang, K.; Wang, W.; Guo, Y.; Wang, X. Prophage encoding toxin/antitoxin system PfiT/PfiA inhibits Pf4 production in Pseudomonas aeruginosa. Microb. Biotechnol. 2020, 13, 1132–1144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zander, I.; Shmidov, E.; Roth, S.; Ben-David, Y.; Shoval, I.; Shoshani, S.; Danielli, A.; Banin, E. Characterization of PfiT/PfiA toxin-antitoxin system of Pseudomonas aeruginosa that affects cell elongation and prophage induction. Environ. Microbiol. 2020, 22, 5048–5057. [Google Scholar] [CrossRef]
- Masuda, Y.; Miyakawa, K.; Nishimura, Y.; Ohtsubo, E. chpA and chpB, Escherichia coli chromosomal homologs of the pem locus responsible for stable maintenance of plasmid R100. J. Bacteriol. 1993, 175, 6850–6856. [Google Scholar] [CrossRef] [Green Version]
- Harms, A.; Maisonneuve, E.; Gerdes, K. Mechanisms of bacterial persistence during stress and antibiotic exposure. Science 2016, 354, aaf4268. [Google Scholar] [CrossRef]
- Lewis, K. Persister cells. Annu. Rev. Microbiol. 2010, 64, 357–372. [Google Scholar] [CrossRef]
- Balaban, N.Q.; Merrin, J.; Chait, R.; Kowalik, L.; Leibler, S. Bacterial persistence as a phenotypic switch. Science 2004, 305, 1622–1625. [Google Scholar] [CrossRef] [Green Version]
- Moyed, H.S.; Bertrand, K.P. hipA, a newly recognized gene of Escherichia coli K-12 that affects frequency of persistence after inhibition of murein synthesis. J. Bacteriol. 1983, 155, 768–775. [Google Scholar] [CrossRef] [Green Version]
- Black, D.S.; Irwin, B.; Moyed, H.S. Autoregulation of hip, an operon that affects lethality due to inhibition of peptidoglycan or DNA synthesis. J. Bacteriol. 1994, 176, 4081–4091. [Google Scholar] [CrossRef] [Green Version]
- Black, D.S.; Kelly, A.J.; Mardis, M.J.; Moyed, H.S. Structure and organization of hip, an operon that affects lethality due to inhibition of peptidoglycan or DNA synthesis. J. Bacteriol. 1991, 173, 5732–5739. [Google Scholar] [CrossRef] [Green Version]
- Korch, S.B.; Hill, T.M. Ectopic overexpression of wild-type and mutant hipA genes in Escherichia coli: Effects on macromolecular synthesis and persister formation. J. Bacteriol. 2006, 188, 3826–3836. [Google Scholar] [CrossRef] [Green Version]
- Schumacher, M.A.; Piro, K.M.; Xu, W.; Hansen, S.; Lewis, K.; Brennan, R.G. Molecular mechanisms of HipA-mediated multidrug tolerance and its neutralization by HipB. Science 2009, 323, 396–401. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Keren, I.; Shah, D.; Spoering, A.; Kaldalu, N.; Lewis, K. Specialized persister cells and the mechanism of multidrug tolerance in Escherichia coli. J. Bacteriol. 2004, 186, 8172–8180. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scherrer, R.; Moyed, H.S. Conditional impairment of cell division and altered lethality in hipA mutants of Escherichia coli K-12. J. Bacteriol. 1988, 170, 3321–3326. [Google Scholar] [CrossRef] [Green Version]
- Wolfson, J.S.; Hooper, D.C.; McHugh, G.L.; Bozza, M.A.; Swartz, M.N. Mutants of Escherichia coli K-12 exhibiting reduced killing by both quinolone and β-lactam antimicrobial agents. Antimicrob. Agents Chemother. 1990, 34, 1938–1943. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maisonneuve, E.; Shakespeare, L.J.; Jorgensen, M.G.; Gerdes, K. Bacterial persistence by RNA endonucleases. Proc. Natl. Acad. Sci. USA 2011, 108, 13206–13211. [Google Scholar] [CrossRef] [Green Version]
- Harms, A.; Fino, C.; Sørensen, M.A.; Semsey, S.; Gerdes, K. Prophages and growth dynamics confound experimental results with antibiotic-tolerant persister cells. mBio 2017, 8, e01964-17. [Google Scholar] [CrossRef] [Green Version]
- Goormaghtigh, F.; Fraikin, N.; Putrinš, M.; Hallaert, T.; Hauryliuk, V.; Garcia-Pino, A.; Sjödin, A.; Kasvandik, S.; Udekwu, K.; Tenson, T.; et al. Reassessing the role of type II toxin-antitoxin systems in formation of Escherichia coli type II persister cells. mBio 2018, 9, e00640-18. [Google Scholar] [CrossRef] [Green Version]
- Ramisetty, B.C.M.; Ghosh, D.; Roy Chowdhury, M.; Santhosh, R.S. What is the link between stringent response, endoribonuclease encoding type ii toxin–antitoxin systems and persistence? Front. Microbiol. 2016, 7, 1882. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tripathi, A.; Dewan, P.C.; Siddique, S.A.; Varadarajan, R. MazF-induced growth inhibition and persister generation in Escherichia coli. J. Biol. Chem. 2014, 289, 4191–4205. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, Y.; Wood, T.K. Toxins Hha and CspD and small RNA regulator Hfq are involved in persister cell formation through MqsR in Escherichia coli. Biochem. Biophys. Res. Commun. 2010, 391, 209–213. [Google Scholar] [CrossRef] [Green Version]
- Luidalepp, H.; Joers, A.; Kaldalu, N.; Tenson, T. Age of inoculum strongly influences persister frequency and can mask effects of mutations implicated in altered persistence. J. Bacteriol. 2011, 193, 3598–3605. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harrison, J.J.; Wade, W.D.; Akierman, S.; Vacchi-Suzzi, C.; Stremick, C.A.; Turner, R.J.; Ceri, H. The chromosomal toxin gene yafQ is a determinant of multidrug tolerance for Escherichia coli growing in a biofilm. Antimicrob. Agents Chemother. 2009, 53, 2253–2258. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amraei, F.; Narimisa, N.; Sadeghi Kalani, B.; Mohammadzadeh, R.; Lohrasbi, V.; Masjedian Jazi, F. The expression of type II TA system genes following exposure to the sub-inhibitory concentration of gentamicin and acid stress in Brucella spp. Microb. Pathog. 2020, 144, 104194. [Google Scholar] [CrossRef]
- Schuster, C.F.; Mechler, L.; Nolle, N.; Krismer, B.; Zelder, M.E.; Gotz, F.; Bertram, R. The MazEF toxin-antitoxin system alters the β-lactam susceptibility of Staphylococcus aureus. PLoS ONE 2015, 10, e0126118. [Google Scholar] [CrossRef] [Green Version]
- Talwar, S.; Pandey, M.; Sharma, C.; Kutum, R.; Lum, J.; Carbajo, D.; Goel, R.; Poidinger, M.; Dash, D.; Singhal, A.; et al. Role of VapBC12 toxin-antitoxin locus in cholesterol-induced mycobacterial persistence. mSystems 2020, 5, e00855-20. [Google Scholar] [CrossRef]
- Diaz-Orejas, R.; Espinosa, M.; Yeo, C.C. The importance of the expendable: Toxin-antitoxin genes in plasmids and chromosomes. Front. Microbiol. 2017, 8, 1479. [Google Scholar] [CrossRef]
- Kirkpatrick, C.L.; Martins, D.; Redder, P.; Frandi, A.; Mignolet, J.; Chapalay, J.B.; Chambon, M.; Turcatti, G.; Viollier, P.H. Growth control switch by a DNA-damage-inducible toxin-antitoxin system in Caulobacter crescentus. Nat. Microbiol. 2016, 1, 16008. [Google Scholar] [CrossRef] [PubMed]
- Qi, Q.; Kamruzzaman, M.; Iredell, J.R. The higBA-type toxin-antitoxin system in IncC plasmids is a mobilizable ciprofloxacin-inducible system. mSphere 2021, e0042421. [Google Scholar] [CrossRef]
- Wang, X.; Wood, T.K. Toxin-antitoxin systems influence biofilm and persister cell formation and the general stress response. Appl. Environ. Microbiol. 2011, 77, 5577–5583. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gottesman, S. Trouble is coming: Signaling pathways that regulate general stress responses in bacteria. J. Biol. Chem. 2019, 294, 11685–11700. [Google Scholar] [CrossRef] [Green Version]
- Narimisa, N.; Amraei, F.; Kalani, B.S.; Mohammadzadeh, R.; Jazi, F.M. Effects of sub-inhibitory concentrations of antibiotics and oxidative stress on the expression of type II toxin-antitoxin system genes in Klebsiella pneumoniae. J. Glob. Antimicrob. Resist. 2020, 21, 51–56. [Google Scholar] [CrossRef]
- Fittipaldi, N.; Segura, M.; Grenier, D.; Gottschalk, M. Virulence factors involved in the pathogenesis of the infection caused by the swine pathogen and zoonotic agent Streptococcus suis. Future Microbiol. 2012, 7, 259–279. [Google Scholar] [CrossRef]
- Forman, H.J.; Torres, M. Reactive oxygen species and cell signaling: Respiratory burst in macrophage signaling. Am. J. Respir. Crit. Care Med. 2002, 166, S4–S8. [Google Scholar] [CrossRef]
- Christensen, S.K.; Mikkelsen, M.; Pedersen, K.; Gerdes, K. RelE, a global inhibitor of translation, is activated during nutritional stress. Proc. Natl. Acad. Sci. USA 2001, 98, 14328–14333. [Google Scholar] [CrossRef] [Green Version]
- Christensen, S.K.; Pedersen, K.; Hansen, F.G.; Gerdes, K. Toxin-antitoxin loci as stress-response-elements: ChpAK/MazF and ChpBK cleave translated RNAs and are counteracted by tmRNA. J. Mol. Biol. 2003, 332, 809–819. [Google Scholar] [CrossRef]
- Tsilibaris, V.; Maenhaut-Michel, G.; Mine, N.; Van Melderen, L. What is the benefit to Escherichia coli of having multiple toxin-antitoxin systems in its genome? J. Bacteriol. 2007, 189, 6101–6108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gerdes, K.; Christensen, S.K.; Lobner-Olesen, A. Prokaryotic toxin-antitoxin stress response loci. Nat. Rev. Microbiol 2005, 3, 371–382. [Google Scholar] [CrossRef] [PubMed]
- Hirakawa, H.; Kodama, T.; Takumi-Kobayashi, A.; Honda, T.; Yamaguchi, A. Secreted indole serves as a signal for expression of type III secretion system translocators in enterohaemorrhagic Escherichia coli O157:H7. Microbiology 2009, 155, 541–550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Masuda, Y.; Sakamoto, E.; Honjoh, K.-I.; Miyamoto, T. Role of toxin-antitoxin-regulated persister population and indole in bacterial heat tolerance. Appl. Environ. Microbiol. 2020, 86, e00935-20. [Google Scholar] [CrossRef]
- Hu, Y.; Kwan, B.W.; Osbourne, D.O.; Benedik, M.J.; Wood, T.K. Toxin YafQ increases persister cell formation by reducing indole signalling. Environ. Microbiol. 2015, 17, 1275–1285. [Google Scholar] [CrossRef]
- Vega, N.M.; Allison, K.R.; Khalil, A.S.; Collins, J.J. Signaling-mediated bacterial persister formation. Nat. Chem. Biol. 2012, 8, 431–433. [Google Scholar] [CrossRef]
- Bleriot, I.; Blasco, L.; Delgado-Valverde, M.; Gual de Torella, A.; Ambroa, A.; Fernandez-Garcia, L.; Lopez, M.; Oteo-Iglesias, J.; Wood, T.K.; Pascual, A.; et al. Mechanisms of tolerance and resistance to chlorhexidine in clinical strains of Klebsiella pneumoniae producers of carbapenemase: Role of new type II toxin-antitoxin system, PemIK. Toxins 2020, 12, 566. [Google Scholar] [CrossRef]
- Stieber, D.; Gabant, P.; Szpirer, C. The art of selective killing: Plasmid toxin/antitoxin systems and their technological applications. Biotechniques 2008, 45, 344–346. [Google Scholar] [CrossRef]
- Pecota, D.C.; Kim, C.S.; Wu, K.; Gerdes, K.; Wood, T.K. Combining the hok/sok, parDE, and pnd postsegregational killer loci to enhance plasmid stability. Appl. Environ. Microbiol. 1997, 63, 1917–1924. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suzuki, M.; Mao, L.; Inouye, M. Single protein production (SPP) system in Escherichia coli. Nat. Protoc. 2007, 2, 1802–1810. [Google Scholar] [CrossRef] [Green Version]
- Suzuki, M.; Zhang, J.; Liu, M.; Woychik, N.A.; Inouye, M. Single protein production in living cells facilitated by an mRNA interferase. Mol. Cell 2005, 18, 253–261. [Google Scholar] [CrossRef]
- Kamruzzaman, M.; Shoma, S.; Thomas, C.M.; Partridge, S.R.; Iredell, J.R. Plasmid interference for curing antibiotic resistance plasmids in vivo. PLoS ONE 2017, 12, e0172913. [Google Scholar] [CrossRef] [PubMed]
- Lazdins, A.; Maurya, A.P.; Miller, C.E.; Kamruzzaman, M.; Liu, S.; Stephens, E.R.; Lloyd, G.S.; Haratianfar, M.; Chamberlain, M.; Haines, A.S.; et al. Potentiation of curing by a broad-host-range self-transmissible vector for displacing resistance plasmids to tackle AMR. PLoS ONE 2020, 15, e0225202. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Pogliano, J.; Helinski, D.R.; Konieczny, I. ParE toxin encoded by the broad-host-range plasmid RK2 is an inhibitor of Escherichia coli gyrase. Mol. Microbiol. 2002, 44, 971–979. [Google Scholar] [CrossRef]
- Pedersen, K.; Zavialov, A.V.; Pavlov, M.Y.; Elf, J.; Gerdes, K.; Ehrenberg, M. The bacterial toxin RelE displays codon-specific cleavage of mRNAs in the ribosomal A site. Cell 2003, 112, 131–140. [Google Scholar] [CrossRef] [Green Version]


| Type II TA | Bacterial Species | Localisation | Function | Reference |
|---|---|---|---|---|
| mvpAT/vapBC | Shigella species | plasmid | maintains virulence plasmid at temperature of human intestine | [50,51] |
| gmvAT | Shigella species | plasmid | maintains virulence plasmid at 21 °C | [50] |
| vapBCST | Salmonella species | plasmid | maintains pSLT virulence plasmid and increases Salmonella survival inside host cell | [9,52,53] |
| ccdABST | Salmonella species | plasmid | maintains pSLT virulence plasmid | [52] |
| pumAB | Pseudomonas aeruginosa | plasmid | facilicates mouse organ invasion and increases C. elegans and mouse mortality rate | [54,55] |
| pemIkSa | Staphylococcus aureus | plasmid | regulation of virulence gene expression | [56] |
| ε/ζ | Streptococcus pneumoniae | chromosome | virulence in mice | [57,58] |
| higBA | P. aeruginosa | chromosome | reduces production of virulence factors | [59] |
| vapBC3, vapBC4 | M. tuberculosis | chromosome | increases pathogenesis in animal model | [60] |
| vapBC11 | M. tuberculosis | chromosome | essential for infection in guinea pigs | [61] |
| mazEF3, mazEF6, mazEF9 | M. tuberculosis | chromosome | increases survival in macrophages and increases colonisation in spleen and lung of guinea pigs | [62] |
| mazEF | S. aureus | chromosome | helps transitioning from acute to chronic infection | [63] |
| vapBC-1,vapXD | Haemophilus influenzae | chromosome | increases survival inside epithelial cells and in the ear of infected chinchillas | [64] |
| toxAvapA | H. influenzae | chromosome | role in chinchilla middle ear infection | [65] |
| ybaJ-hha, yefM-yoeB | E. coli | chromosome | contributes to the colonisation in the bladder | [66] |
| pasTI | E. coli | chromosome | contributes to the colonisation in the kidneys | [66] |
| relBE4,relBE7 | V. cholerae | chromosome II | improves intestinal colonisation in mice | [67] |
| fitBA | Neisseria gonorrhoeae | chromosome | intracellular growth regulator | [68] |
| pezAT | S. pneumoniae | chromosome | virulence in mice | [58,69] |
| higBA | P. aeruginosa | chromosome | reduces virulence factors production | [59] |
| SehAB | Salmonella enterica | chromosome | reduces virulence in mice | [49] |
| savRS | S. aureus | chromosome | negatively regulates the virulence gene expression and pathogenicity | [70] |
| higBA | P. aeruginosa | chromosome | inhibits virulence gene expression | [71] |
| Rhs locus | Salmonella Typhimurium | chromosome | represses proliferation within host macrophages | [72,73,74,75] |
| Type II TA | Bacterial Species | Localisation | Function | Reference |
|---|---|---|---|---|
| parDE | E. coli | plasmid | promotes biofilm formation | [8] |
| mqsRA | E. coli | chromosome | biofilm production/controversial | [104] |
| mazEF and dinJ-yqfQ | E. coli | chromosome | promotes biofilm production | [105] |
| hipAB | Shewanella oneidensis and E. coli. | chromosome | promotes biofilm production | [106] |
| yqcGF | Bacillus subtilis | chromosome | promotes biofilm production | [107] |
| Rv2871-Rv2872 | M. tuberculosis | chromosome | enhances biofilm development | [108] |
| higBA | P. aeruginosa | chromosome | repression of biofilm production | [109] |
| yefM-yoeB, RelBE | S. pneumoniae | chromosome | promotes biofilm formation | [110] |
| relBE and variants | V. cholerae | chromosome | promotes biofilm formation and biofilm maturation | [67] |
| higBA | P. aeruginosa | chromosome | reduces biofilm formation | [59] |
| mazEF | S. aureus | chromosome | promotes biofilm formation | [63] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kamruzzaman, M.; Wu, A.Y.; Iredell, J.R. Biological Functions of Type II Toxin-Antitoxin Systems in Bacteria. Microorganisms 2021, 9, 1276. https://doi.org/10.3390/microorganisms9061276
Kamruzzaman M, Wu AY, Iredell JR. Biological Functions of Type II Toxin-Antitoxin Systems in Bacteria. Microorganisms. 2021; 9(6):1276. https://doi.org/10.3390/microorganisms9061276
Chicago/Turabian StyleKamruzzaman, Muhammad, Alma Y. Wu, and Jonathan R. Iredell. 2021. "Biological Functions of Type II Toxin-Antitoxin Systems in Bacteria" Microorganisms 9, no. 6: 1276. https://doi.org/10.3390/microorganisms9061276







