Conversion of the Sensor Kinase DcuS to the Fumarate Sensitive State by Interaction of the Bifunctional Transporter DctA at the TM2/PASC-Linker Region
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Growth Conditions
2.2. BACTH and β-galactosidaseAassay
2.3. Time-Resolved Oxidative Cys-Cross-Linking
2.4. In Silico Analysis
3. Results
3.1. The TM2/PASC-Linker of DcuS Plays an Important Role for the Interaction with H8b of DctA
3.2. Absence of DctA Affects Cys-Cross-Linking Efficiency in the TM2/PASC-Linker of DcuS
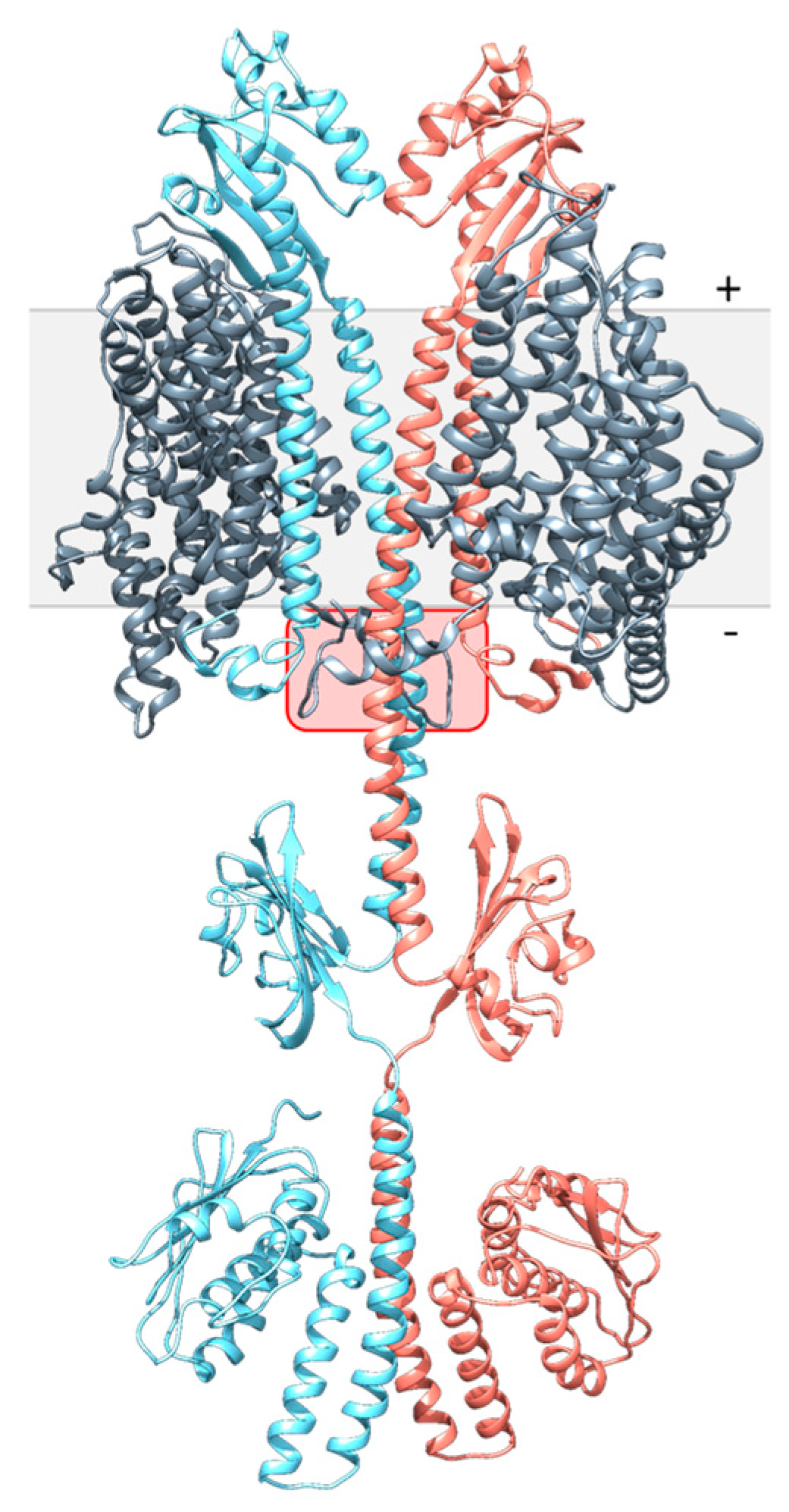
3.3. Modelling of DcuS and DctA
3.4. Structural Modelling of the DcuS Linker and DctA H8b Interaction
4. Discussion
4.1. DctA as a Bifunctional Protein: Function as a Transporter and as a Structural Co-Regulator of DcuS
DctA as a Transporter for C4DCs
4.2. DctA as a Structural Co-Regulator of DcuS and Formation of the DctA × DcuS Sensor Complex
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mascher, T.; Helmann, J.D.; Unden, G. Stimulus perception in bacterial signal-transducing histidine kinases. Microbiol. Mol. Biol. Rev. 2006, 70, 910–938. [Google Scholar] [CrossRef] [PubMed]
- Krell, T.; Lacal, J.; Busch, A.; Silva-Jiménez, H.; Guazzaroni, M.-E.; Ramos, J.L. Bacterial sensor kinases: Diversity in the recognition of environmental signals. Annu. Rev. Microbiol. 2010, 64, 539–559. [Google Scholar] [CrossRef] [PubMed]
- Zientz, E.; Bongaerts, J.; Unden, G. Fumarate regulation of gene expression in Escherichia coli by the DcuSR (dcuSR genes) two-component regulatory system. J. Bacteriol. 1998, 180, 5421–5425. [Google Scholar] [CrossRef]
- Janausch, I.G.; Zientz, E.; Tran, Q.H.; Kröger, A.; Unden, G. C4-dicarboxylate carriers and sensors in bacteria. BBA Bioenerg. 2002, 1553, 39–56. [Google Scholar] [CrossRef]
- Unden, G.; Strecker, A.; Kleefeld, A.; Kim, O.B. C4-dicarboxylate utilization in aerobic and anaerobic growth. EcoSal Plus 2016, 7. [Google Scholar] [CrossRef]
- Scheu, P.D.; Liao, Y.-F.; Bauer, J.; Kneuper, H.; Basche, T.; Unden, G.; Erker, W. Oligomeric sensor kinase DcuS in the membrane of Escherichia coli and in proteoliposomes: Chemical cross-linking and FRET spectroscopy. J. Bacteriol. 2010, 192, 3474–3483. [Google Scholar] [CrossRef] [PubMed]
- Pappalardo, L.; Janausch, I.G.; Vijayan, V.; Zientz, E.; Junker, J.; Peti, W.; Zweckstetter, M.; Unden, G.; Griesinger, C. The NMR structure of the sensory domain of the membranous two-component fumarate sensor (histidine protein kinase) DcuS of Escherichia coli. J. Biol. Chem. 2003, 278, 39185–39188. [Google Scholar] [CrossRef]
- Kneuper, H.; Janausch, I.G.; Vijayan, V.; Zweckstetter, M.; Bock, V.; Griesinger, C.; Unden, G. The nature of the stimulus and of the fumarate binding site of the fumarate sensor DcuS of Escherichia coli. J. Biol. Chem. 2005, 280, 20596–20603. [Google Scholar] [CrossRef]
- Cheung, J.; Hendrickson, W.A. Crystal structure of the E. coli DcuS sensor domain. J. Biol. Chem. 2008, 283, 13762–13770. [Google Scholar] [CrossRef]
- Sevvana, M.; Vijayan, V.; Zweckstetter, M.; Reinelt, S.; Madden, D.R.; Herbst-Irmer, R.; Sheldrick, G.M.; Bott, M.; Griesinger, C.; Becker, S. A ligand-induced switch in the periplasmic domain of sensor histidine kinase CitA. J. Mol. Biol. 2008, 377, 512–523. [Google Scholar] [CrossRef]
- Salvi, M.; Schomburg, B.; Giller, K.; Graf, S.; Unden, G.; Becker, S.; Lange, A.; Griesinger, C. Sensory domain contraction in histidine kinase CitA triggers transmembrane signaling in the membrane-bound sensor. Proc. Natl. Acad. Sci. USA 2017, 114, 3115–3120. [Google Scholar] [CrossRef]
- Monzel, C.; Unden, G. Transmembrane signaling in the sensor kinase DcuS of Escherichia coli: A long-range piston-type displacement of transmembrane helix 2. Proc. Natl. Acad. Sci. USA 2015, 112, 11042–11047. [Google Scholar] [CrossRef]
- Stopp, M.; Steinmetz, P.A.; Schubert, C.; Griesinger, C.; Schneider, D.; Unden, G. Transmembrane signaling and cytoplasmic signal conversion by dimeric transmembrane helix 2 and a linker domain of the DcuS sensor kinase. J. Biol. Chem. 2021, 296, 100148. [Google Scholar] [CrossRef]
- Davies, S.J.; Golby, P.; Omrani, D.; Broad, S.A.; Harrington, V.L.; Guest, J.R.; Kelly, D.J.; Andrews, S.C. Inactivation and regulation of the aerobic C4-dicarboxylate transport (dctA) gene of Escherichia coli. J. Bacteriol. 1999, 181, 5624–5635. [Google Scholar] [CrossRef]
- Kleefeld, A.; Ackermann, B.; Bauer, J.; Krämer, J.; Unden, G. The fumarate/succinate antiporter DcuB of Escherichia coli is a bifunctional protein with sites for regulation of DcuS dependent gene expression. J. Biol. Chem. 2009, 284, 265–275. [Google Scholar] [CrossRef]
- Witan, J.; Bauer, J.; Wittig, I.; Steinmetz, P.A.; Erker, W.; Unden, G. Interaction of the Escherichia coli transporter DctA with the sensor kinase DcuS: Presence of functional DctA/DcuS sensor units. Mol. Microbiol. 2012, 85, 846–861. [Google Scholar] [CrossRef]
- Steinmetz, P.A.; Worner, S.; Unden, G. Differentiation of DctA and DcuS function in the DctA/DcuS sensor complex of Escherichia coli: Function of DctA as an activity switch and of DcuS as the C4-dicarboxylate sensor. Mol. Microbiol. 2014, 94, 218–229. [Google Scholar] [CrossRef] [PubMed]
- Wörner, S.; Strecker, A.; Monzel, C.; Zeltner, M.; Witan, J.; Ebert-Jung, A.; Unden, G. Conversion of the sensor kinase DcuS of Escherichia coli of the DcuB/DcuS sensor complex to the C4-dicarboxylate responsive form by the transporter DcuB. Environ. Microbiol. 2016, 18, 4920–4930. [Google Scholar] [CrossRef] [PubMed]
- Engel, P.; Krämer, R.; Unden, G. Transport of C4-dicarboxylates by anaerobically grown Escherichia coli. Eur J Biochem. 1994, 222, 605–614. [Google Scholar] [CrossRef] [PubMed]
- Six, S.; Andrews, S.C.; Unden, G.; Guest, J.R. Escherichia coli possesses two homologous anaerobic C4-dicarboxylate membrane transporters (DcuA and DcuB) distinct from the aerobic dicarboxylate transport system (Dct). J. Bacteriol. 1994, 176, 6470–6478. [Google Scholar] [CrossRef] [PubMed]
- Witan, J.; Monzel, C.; Scheu, P.D.; Unden, G. The sensor kinase DcuS of Escherichia coli: Two stimulus input sites and a merged signal pathway in the DctA/DcuS sensor unit. Biol. Chem. 2012, 393, 1291–1297. [Google Scholar] [CrossRef]
- Miller, J.H. A Short Course in Bacterial Genetics: A Laboratory Manual and Handbook for Escherichia coli and Related Bacteria; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 1992; ISBN 0879693495. [Google Scholar]
- Jones, H.M.; Gunsalus, R.P. Regulation of Escherichia coli fumarate reductase (frdABCD) operon expression by respiratory electron acceptors and the fnr gene product. J. Bacteriol. 1987, 169, 3340–3349. [Google Scholar] [CrossRef]
- Monzel, C.; Degreif-Dünnwald, P.; Gröpper, C.; Griesinger, C.; Unden, G. The cytoplasmic PASC domain of the sensor kinase DcuS of Escherichia coli: Role in signal transduction, dimer formation, and DctA interaction. Microbiol. Open 2013, 2, 912–927. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular Cloning: A Laboratory Manual, 2nd ed.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 1989. [Google Scholar]
- Weisenburger, S.; Boening, D.; Schomburg, B.; Giller, K.; Becker, S.; Griesinger, C.; Sandoghdar, V. Cryogenic optical localization provides 3D protein structure data with Angstrom resolution. Nat. Methods 2017, 14, 141–144. [Google Scholar] [CrossRef] [PubMed]
- Etzkorn, M.; Kneuper, H.; Dünnwald, P.; Vijayan, V.; Krämer, J.; Griesinger, C.; Becker, S.; Unden, G.; Baldus, M. Plasticity of the PAS domain and a potential role for signal transduction in the histidine kinase DcuS. Nat. Struct. Mol. Biol. 2008, 15, 1031. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Zhang, Y. I-TASSER server: New development for protein structure and function predictions. Nucleic Acids Res. 2015, 43, W174–W181. [Google Scholar] [CrossRef] [PubMed]
- Podgornaia, A.I.; Casino, P.; Marina, A.; Laub, M.T. Structural basis of a rationally rewired protein-protein interface critical to bacterial signaling. Structure 2013, 21, 1636–1647. [Google Scholar] [CrossRef]
- UniProt Consortium. UniProt: A worldwide hub of protein knowledge. Nucleic Acids Res. 2019, 47, D506–D515. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
- Golby, P.; Kelly, D.J.; Guest, J.R.; Andrews, S.C. Transcriptional regulation and organization of the dcuA and dcuB genes, encoding homologous anaerobic C4-dicarboxylate transporters in Escherichia coli. J. Bacteriol. 1998, 180, 6586–6596. [Google Scholar] [CrossRef]
- Lee, G.F.; Lebert, M.R.; Lilly, A.A.; Hazelbauer, G.L. Transmembrane signaling characterized in bacterial chemoreceptors by using sulfhydryl cross-linking in vivo. Proc. Natl. Acad. Sci. USA 1995, 92, 3391–3395. [Google Scholar] [CrossRef]
- Jensen, S.; Guskov, A.; Rempel, S.; Hänelt, I.; Slotboom, D.J. Crystal structure of a substrate-free aspartate transporter. Nat. Struct. Mol. Biol. 2013, 20, 1224–1226. [Google Scholar] [CrossRef] [PubMed]
- Bordoli, L.; Kiefer, F.; Arnold, K.; Benkert, P.; Battey, J.; Schwede, T. Protein structure homology modeling using SWISS-MODEL workspace. Nat. Protoc. 2009, 4, 1–13. [Google Scholar] [CrossRef]
- Scheu, P.D.; Witan, J.; Rauschmeier, M.; Graf, S.; Liao, Y.-F.; Ebert-Jung, A.; Basche, T.; Erker, W.; Unden, G. CitA/CitB two-component system regulating citrate fermentation in Escherichia coli and its relation to the DcuS/DcuR system in vivo. J. Bacteriol. 2012, 194, 636–645. [Google Scholar] [CrossRef] [PubMed]
- Okonechnikov, K.; Golosova, O.; Fursov, M. Unipro UGENE: A unified bioinformatics toolkit. Bioinformatics 2012, 28, 1166–1167. [Google Scholar] [CrossRef]
- Von Heijne, G. Membrane-protein topology. Nat. Rev. Mol. Cell Biol. 2006, 7, 909–918. [Google Scholar] [CrossRef] [PubMed]
- Kay, W.W.; Kornberg, H.L. The uptake of C4-dicarboxylic acids by Escherichia coli. FEBS 1971, 18, 274–281. [Google Scholar] [CrossRef]
- Wörner, S.; Surmann, K.; Ebert-Jung, A.; Völker, U.; Hammer, E.; Unden, G. Cellular concentrations of the transporters DctA, DcuB, and the sensor DcuS of Escherichia coli and the contributions of free and complexed DcuS to transcriptional regulation by DcuR. J. Bacteriol. 2017, 200, e00612-17. [Google Scholar] [CrossRef]
- Scheu, P.; Sdorra, S.; Liao, Y.-F.; Wegner, M.; Basche, T.; Unden, G.; Erker, W. Polar accumulation of the metabolic sensory histidine kinases DcuS and CitA in Escherichia coli. Microbiology 2008, 154, 2463–2472. [Google Scholar] [CrossRef][Green Version]
- Scheu, P.D.; Steinmetz, P.A.; Dempwolff, F.; Graumann, P.L.; Unden, G. Polar localization of a tripartite complex of the two-component system DcuS/DcuR and the transporter DctA in Escherichia coli depends on the sensor kinase DcuS. PLoS ONE 2014, 9, e115534. [Google Scholar] [CrossRef] [PubMed]
- Strecker, A.; Schubert, C.; Zedler, S.; Steinmetz, P.; Unden, G. DcuA of aerobically grown Escherichia coli serves as a nitrogen shuttle (L-aspartate/fumarate) for nitrogen uptake. Mol. Microbiol. 2018, 109, 801–811. [Google Scholar] [CrossRef]
- Krämer, J.; Fischer, J.D.; Zientz, E.; Vijayan, V.; Griesinger, C.; Lupas, A.; Unden, G. Citrate sensing by the C4-dicarboxylate/citrate sensor kinase DcuS of Escherichia coli: Binding site and conversion of DcuS to a C4-dicarboxylate-or citrate-specific sensor. J. Bacteriol. 2007, 189, 4290–4298. [Google Scholar] [CrossRef] [PubMed]
- Piepenbreier, H.; Fritz, G.; Gebhard, S. Transporters as information processors in bacterial signalling pathways. Mol. Microbiol. 2017, 104, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Karimova, G.; Pidoux, J.; Ullmann, A.; Ladant, D. A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc. Natl. Acad. Sci. USA 1998, 95, 5752–5756. [Google Scholar] [CrossRef]
- Miroux, B.; Walker, J.E. Over-production of proteins in Escherichia coli: Mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J. Mol. Biol. 1996, 260, 289–298. [Google Scholar] [CrossRef]
- Baba, T.; Ara, T.; Hasegawa, M.; Takai, Y.; Okumura, Y.; Baba, M.; Datsenko, K.A.; Tomita, M.; Wanner, B.L.; Mori, H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: The Keio collection. Mol. Syst. Biol. 2006, 2, 2006-0008. [Google Scholar] [CrossRef]
- Karimova, G.; Ullmann, A.; Ladant, D. Protein-protein interaction between Bacillus stearothermophilus tyrosyl-tRNA synthetase subdomains revealed by a bacterial two-hybrid system. J. Mol. Microbiol. Biotechnol. 2001, 3, 73–82. [Google Scholar]





| Cys Cross-Linking Increase (% of CL per min) | ||||
|---|---|---|---|---|
| DcuS variant | E. coli C43(DE3) (Wt) | E. coli IMW660 (DctA−) | ||
| − Fum | + Fum | − Fum | + Fum | |
| G190C (TM2) | 9.2 | 9.7 | 7.7 | 6.5 |
| G194C (TM2) | 7.8 | 7.1 | 7.3 | 7.7 |
| I208C (Linker) | 2.6 | -0.1 | 0.0 | 0.0 |
| L209C (Linker) | 2.8 | 1.4 | 0.0 | 0.0 |
| R224C (PASc) | 3.5 | 2.7 | 2.1 | 1.0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stopp, M.; Schubert, C.; Unden, G. Conversion of the Sensor Kinase DcuS to the Fumarate Sensitive State by Interaction of the Bifunctional Transporter DctA at the TM2/PASC-Linker Region. Microorganisms 2021, 9, 1397. https://doi.org/10.3390/microorganisms9071397
Stopp M, Schubert C, Unden G. Conversion of the Sensor Kinase DcuS to the Fumarate Sensitive State by Interaction of the Bifunctional Transporter DctA at the TM2/PASC-Linker Region. Microorganisms. 2021; 9(7):1397. https://doi.org/10.3390/microorganisms9071397
Chicago/Turabian StyleStopp, Marius, Christopher Schubert, and Gottfried Unden. 2021. "Conversion of the Sensor Kinase DcuS to the Fumarate Sensitive State by Interaction of the Bifunctional Transporter DctA at the TM2/PASC-Linker Region" Microorganisms 9, no. 7: 1397. https://doi.org/10.3390/microorganisms9071397
APA StyleStopp, M., Schubert, C., & Unden, G. (2021). Conversion of the Sensor Kinase DcuS to the Fumarate Sensitive State by Interaction of the Bifunctional Transporter DctA at the TM2/PASC-Linker Region. Microorganisms, 9(7), 1397. https://doi.org/10.3390/microorganisms9071397






