Enterocin Cross-Resistance Mediated by ABC Transport Systems
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Culture Media
2.2. Inhibitory-Activity Assays
2.3. Bacteriocin Production and Purification
2.4. mr10A/B Gene Cluster Sequencing and Annotation, and Genetic Data
2.5. Cluster Analyses
3. Results
3.1. mr10A/B Gene Cluster
3.2. Homologies between the ABC Transporters
3.3. Mutant Sensitivity to Enterocins MR10A/B
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ghoul, M.; Mitri, S. The Ecology and Evolution of Microbial Competition. Trends Microbiol. 2016, 24, 833–845. [Google Scholar] [CrossRef]
- Dobson, A.; Cotter, P.D.; Ross, R.P.; Hill, C. Bacteriocin Production: A Probiotic Trait? Appl. Environ. Microbiol. 2012, 78, 1–6. [Google Scholar] [CrossRef]
- Wang, H.; Fewer, D.P.; Sivonen, K. Genome Mining Demonstrates the Widespread Occurrence of Gene Clusters Encoding Bacteriocins in Cyanobacteria. PLoS ONE 2011, 6, e22384. [Google Scholar] [CrossRef]
- Rebuffat, S. Bacteriocins from Gram-Negative Bacteria: A Classification? Prokaryotic Antimicrob. Pept. 2011, 55–72. [Google Scholar] [CrossRef]
- Zacharof, M.P.; Lovitt, R.W. Bacteriocins Produced by Lactic Acid Bacteria a Review Article. APCBEE Procedia 2012, 2, 50–56. [Google Scholar] [CrossRef]
- Kommineni, S.; Bretl, D.J.; Lam, V.; Chakraborty, R.; Hayward, M.; Simpson, P.; Cao, Y.; Bousounis, P.; Kristich, C.J.; Salzman, N.H. Bacteriocin Production Augments Niche Competition by Enterococci in the Mammalian Gastrointestinal Tract. Nature 2015, 526, 719–722. [Google Scholar] [CrossRef] [PubMed]
- Soltani, S.; Hammami, R.; Cotter, P.D.; Rebuffat, S.; Said, L.B.; Gaudreau, H.; Bédard, F.; Biron, E.; Drider, D.; Fliss, I. Bacteriocins as a New Generation of Antimicrobials: Toxicity Aspects and Regulations. FEMS Microbiol. Rev. 2020, 39, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Meade, E.; Slattery, M.A.; Garvey, M. Bacteriocins, Potent Antimicrobial Peptides and the Fight against Multi Drug Resistant Species: Resistance Is Futile? Antibiotics 2020, 9, 32. [Google Scholar] [CrossRef]
- Liu, G.; Song, Z.; Yang, X.; Gao, Y.; Wang, C.; Sun, B. Antibacterial Mechanism of Bifidocin A, a Novel Broad-Spectrum Bacteriocin Produced by Bifidobacterium Animalis BB04. Food Control 2016, 62, 309–316. [Google Scholar] [CrossRef]
- Fallico, V.; McAuliffe, O.; Ross, R.P.; Fitzgerald, G.F.; Hill, C. The potential of lacticin 3147, enterocin AS-48, lacticin 481, variacin and sakacin P for food biopreservation. In Protective Cultures, Antimicrobial Metabolites and Bacteriophages for Food and Beverage Biopreservation; Elsevier: Amsterdam, The Netherlands, 2011; pp. 100–121. ISBN 978-1-84569-669-6. [Google Scholar]
- Jozala, A.F.; Novaes, L.C.d.L.; Júnior, A.P. Nisin. In Concepts, Compounds and the Alternatives of Antibacterials; IntechOpen: London, UK, 2015; pp. 103–119. [Google Scholar] [CrossRef]
- Alvarez-Sieiro, P.; Montalbán-López, M.; Mu, D.; Kuipers, O.P. Bacteriocins of Lactic Acid Bacteria: Extending the Family. Appl. Microbiol. Biotechnol. 2016, 100, 2939–2951. [Google Scholar] [CrossRef]
- Kumariya, R.; Garsa, A.K.; Rajput, Y.S.; Sood, S.K.; Akhtar, N.; Patel, S. Bacteriocins: Classification, Synthesis, Mechanism of Action and Resistance Development in Food Spoilage Causing Bacteria. Microb. Pathog. 2019, 128, 171–177. [Google Scholar] [CrossRef] [PubMed]
- Cintas, L.M.; Casaus, M.P.; Herranz, C.; Nes, I.F.; Hernández, P.E. Review: Bacteriocins of Lactic Acid Bacteria. Food Sci. Technol. Int. 2001, 7, 281–305. [Google Scholar] [CrossRef]
- Dimov, S.; Ivanova, P.; Harizanova, N. Genetics of Bacteriocins Biosynthesis by Lactic Acid Bacteria. Biotechnol. Biotechnol. Equip. 2005, 193, 4–10. [Google Scholar] [CrossRef]
- de Freire Bastos, M.D.C.; Varella Coelho, M.L.; da Silva Santos, O.C. Resistance to Bacteriocins Produced by Gram-Positive Bacteria. Microbiology 2015, 161, 683–700. [Google Scholar] [CrossRef]
- Maqueda, M.; Galvez, A.; Bueno, M.; Sanchez-Barrena, M.; Gonzalez, C.; Albert, A.; Rico, M.; Valdivia, E. Peptide AS-48: Prototype of a New Class of Cyclic Bacteriocins. Curr. Protein Pept. Sci. 2004, 5, 399–416. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Bueno, M.; Galvez, A.; Valdivia, E.; Maqueda, M. A Transferable Plasmid Associated with AS-48 Production in Enterococcus Faecalis. J. Bacteriol. 1990, 172, 2817–2818. [Google Scholar] [CrossRef]
- Cebrián, R.; Rodríguez-Ruano, S.; Martínez-Bueno, M.; Valdivia, E.; Maqueda, M.; Montalbán-López, M. Analysis of the Promoters Involved in Enterocin AS-48 Expression. PLoS ONE 2014, 9, e90603. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Bueno, M.; Valdivia, E.; Gálvez, A.; Coyette, J.; Maqueda, M. Analysis of the Gene Cluster Involved in Production and Immunity of the Peptide Antibiotic AS-48 in Enterococcus Faecalis. Mol. Microbiol. 1998, 27, 347–358. [Google Scholar] [CrossRef]
- Diaz, M.; Valdivia, E.; Martínez-Bueno, M.; Fernández, M.; Santos Soler-González, A.; Ramírez-Rodrigo, H.; Maqueda, M. Characterization of a New Operon, as-48EFGH, from the as-48 Gene Cluster Involved in Immunity to Enterocin AS-48. Appl. Environ. Microbiol. 2003, 69, 1229–1236. [Google Scholar] [CrossRef]
- Du Toit, M.; Franz, C.M.A.P.; Dicks, L.M.T.; Holzapfel, W.H. Preliminary Characterization of Bacteriocins Produced by Enterococcus Faecium and Enterococcus Faecalis Isolated from Pig Faeces. J. Appl. Microbiol. 2000, 88, 482–494. [Google Scholar] [CrossRef] [PubMed]
- De Vuyst, L.; Foulquié Moreno, M.R.; Revets, H. Screening for Enterocins and Detection of Hemolysin and Vancomycin Resistance in Enterococci of Different Origins. Int. J. Food Microbiol. 2003, 84, 299–318. [Google Scholar] [CrossRef]
- Guerra, B.; Junker, E.; Schroeter, A.; Malorny, B.; Lehmann, S.; Helmuth, R. Phenotypic and Genotypic Characterization of Antimicrobial Resistance in German Escherichia Coli Isolates from Cattle, Swine and Poultry. J. Antimicrob. Chemother. 2003, 52, 489–492. [Google Scholar] [CrossRef]
- Ogaki, M.B.; Rocha, K.R.; Terra, M.R.; Furlaneto, M.C.; Furlaneto-Maia, L. Screening of the Enterocin-Encoding Genes and Antimicrobial Activity in Enterococcus Species. J. Microbiol. Biotechnol. 2016, 26, 1026–1034. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Barba, J.L.; Floriano, B.; Maldonado-Barragán, A.; Jiménez-Díaz, R. Molecular Analysis of the 21-Kb Bacteriocin-Encoding Plasmid PEF1 from Enterococcus Faecium 6T1a. Plasmid 2007, 57, 175–181. [Google Scholar] [CrossRef] [PubMed]
- Achemchem, F.; Martínez-Bueno, M.; Abrini, J.; Valdivia, E.; Maqueda, M. Enterococcus Faecium F58, a Bacteriocinogenic Strain Naturally Occurring in Jben, a Soft, Farmhouse Goat’s Cheese Made in Morocco. J. Appl. Microbiol. 2005, 99, 141–150. [Google Scholar] [CrossRef]
- Martín-Platero, A.M.; Valdivia, E.; Ruiz-Rodríguez, M.; Soler, J.J.; Martín-Vivaldi, M.; Maqueda, M.; Martínez-Bueno, M. Characterization of Antimicrobial Substances Produced by Enterococcus Faecalis MRR 10-3, Isolated from the Uropygial Gland of the Hoopoe (Upupa Epops). Appl. Environ. Microbiol. 2006, 72, 4245–4249. [Google Scholar] [CrossRef]
- Chapman, J.S. Disinfectant Resistance Mechanisms, Cross-Resistance, and Co-Resistance. Int. Biodeterior. Biodegrad. 2003, 51, 271–276. [Google Scholar] [CrossRef]
- Gravesen, A.; Ramnath, M.; Rechinger, K.B.; Andersen, N.; Jänsch, L.; Héchard, Y.; Hastings, J.W.; Knøchel, S. High-Level Resistance to Class IIa Bacteriocins Is Associated with One General Mechanism in Listeria Monocytogenes. Microbiology 2002, 148, 2361–2369. [Google Scholar] [CrossRef]
- Vignolo, G.; Palacios, J.; Farías, M.E.; Sesma, F.; Schillinger, U.; Holzapfel, W.; Oliver, G. Combined Effect of Bacteriocins on the Survival of Various Listeria Species in Broth and Meat System. Curr. Microbiol. 2000, 41, 410–416. [Google Scholar] [CrossRef]
- Kaur, G.; Singh, T.P.; Malik, R.K. Antibacterial Efficacy of Nisin, Pediocin 34 and Enterocin FH99 against Listeria Monocytogenes and Cross Resistance of Its Bacteriocin Resistant Variants to Common Food Preservatives. Braz. J. Microbiol. 2013, 44, 63–71. [Google Scholar] [CrossRef]
- Fimland, G.; Eijsink, V.G.H.; Nissen-Meyer, J. Comparative Studies of Immunity Proteins of Pediocin-like Bacteriocins. Microbiology 2002, 148, 3661–3670. [Google Scholar] [CrossRef][Green Version]
- Oppegård, C.; Emanuelsen, L.; Thorbek, L.; Fimland, G.; Nissen-Meyer, J. The Lactococcin G Immunity Protein Recognizes Specific Regions in Both Peptides Constituting the Two-Peptide Bacteriocin Lactococcin G. Appl. Environ. Microbiol 2010, 76, 1267–1273. [Google Scholar] [CrossRef] [PubMed]
- Fernández, M.; Sánchez-Hidalgo, M.; García-Quintáns, N.; Martínez-Bueno, M.; Valdivia, E.; López, P.; Maqueda, M. Processing of As-48ABC RNA in AS-48 Enterocin Production by Enterococcus Faecalis. J. Bacteriol. 2008, 190, 240–250. [Google Scholar] [CrossRef] [PubMed]
- Achemchem, F.; Abrini, J.; Martínez-Bueno, M.; Valdivia, E.; Maqueda, M. Control of Listeria Monocytogenes in Goat’s Milk and Goat’s Jben by the Bacteriocinogenic Enterococcus Faecium F58 Strain. J. Food Prot. 2006, 69, 2370–2376. [Google Scholar] [CrossRef] [PubMed]
- Wirth, R.; An, F.Y.; Clewell, D.B. Highly Efficient Protoplast Transformation System for Streptococcus Faecalis and a New Escherichia Coli-S. Faecalis Shuttle Vector. J. Bacteriol. 1986, 165, 831–836. [Google Scholar] [CrossRef] [PubMed]
- Galvez, A.; Valdivia, E.; Maqueda, M.; Montoya, E. Production of Bacteriocin-like Substances by Group D Streptococci of Human Origin. Microbios 1985, 43, 223–232. [Google Scholar]
- Balouiri, M.; Sadiki, M.; Ibnsouda, S.K. Methods for in Vitro Evaluating Antimicrobial Activity: A Review. J. Pharm. Anal. 2016, 6, 71–79. [Google Scholar] [CrossRef]
- Abriouel, H.; Maqueda, M.; Gálvez, A.; Martínez-Bueno, M.; Valdivia, E. Inhibition of Bacterial Growth, Enterotoxin Production, and Spore Outgrowth in Strains of Bacillus Cereus by Bacteriocin AS-48. Appl. Environ. Microbiol. 2002, 68, 1473–1477. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Seemann, T. Prokka: Rapid Prokaryotic Genome Annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Zhang, Z.; Schwartz, S.; Wagner, L.; Miller, W. A Greedy Algorithm for Aligning DNA Sequences. J. Comput. Biol. 2000, 7, 203–214. [Google Scholar] [CrossRef]
- Altschul, S.F.; Wootton, J.C.; Gertz, E.M.; Agarwala, R.; Morgulis, A.; Schäffer, A.A.; Yu, Y.-K. Protein Database Searches Using Compositionally Adjusted Substitution Matrices. FEBS J. 2005, 272, 5101–5109. [Google Scholar] [CrossRef]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A New Generation of Protein Database Search Programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2020. [Google Scholar]
- RStudio Team. RStudio: Integrated Development Environment for R; RStudio, Inc.: Boston, MA, USA, 2016. [Google Scholar]
- Gu, Z.; Eils, R.; Schlesner, M. Complex Heatmaps Reveal Patterns and Correlations in Multidimensional Genomic Data. Bioinformatics 2016, 32, 2847–2849. [Google Scholar] [CrossRef]
- Garnier, S. Viridis: Default Color Maps from “Matplotlib”, R package version 0.5 1; R Foundation for Statistical Computing: Vienna, Austria, 2018. [Google Scholar]
- Galili, T. Dendextend: An R Package for Visualizing, Adjusting and Comparing Trees of Hierarchical Clustering. Bioinformatics 2015, 31, 3718–3720. [Google Scholar] [CrossRef] [PubMed]
- Todorov, S.D.; Franco, B.D.G.d.M.; Tagg, J.R. Bacteriocins of Gram-Positive Bacteria Having Activity Spectra Extending beyond Closely-Related Species. Benef. Microbes 2019, 10, 315–328. [Google Scholar] [CrossRef] [PubMed]
- Cotter, P.D.; Ross, R.P.; Hill, C. Bacteriocins—A Viable Alternative to Antibiotics? Nat. Rev. Microbiol. 2013, 11, 95–105. [Google Scholar] [CrossRef]
- Reygaert, W.C. An Overview of the Antimicrobial Resistance Mechanisms of Bacteria. AIMS Microbiol. 2018, 4, 482–501. [Google Scholar] [CrossRef]
- Tenover, F.C. Mechanisms of Antimicrobial Resistance in Bacteria. Am. J. Med. 2006, 119, S3–S10. [Google Scholar] [CrossRef]
- Towle, K.M.; Vederas, J.C. Structural Features of Many Circular and Leaderless Bacteriocins Are Similar to Those in Saposins and Saposin-like Peptides. Med. Chem. Commun. 2017, 8, 276–285. [Google Scholar] [CrossRef]
- Ventola, C.L. The Antibiotic Resistance Crisis. P T 2015, 40, 277–283. [Google Scholar] [PubMed]
- WHO. Global Action Plan on Antimicrobial Resistance. Available online: https://www.who.int/publications-detail-redirect/global-action-plan-on-antimicrobial-resistance (accessed on 8 January 2021).
- Lei, J.; Sun, L.C.; Huang, S.; Zhu, C.; Li, P.; He, J.; Mackey, V.; Coy, D.H.; He, Q.Y. The Antimicrobial Peptides and Their Potential Clinical Applications. Am. J. Transl. Res. 2019, 11, 3919–3931. [Google Scholar] [PubMed]
- Mahlapuu, M.; Håkansson, J.; Ringstad, L.; Björn, C. Antimicrobial Peptides: An Emerging Category of Therapeutic Agents. Front. Cell. Infect. Microbiol. 2016, 6, 194. [Google Scholar] [CrossRef] [PubMed]
- Saleh-Mghir, A.; Muller-Serieys, C.; Dinh, A.; Massias, L.; Crémieux, A.C. Adjunctive Rifampin Is Crucial to Optimizing Daptomycin Efficacy against Rabbit Prosthetic Joint Infection Due to Methicillin-Resistant Staphylococcus Aureus. Antimicrob. Agents Chemother. 2011, 55, 4589–4593. [Google Scholar] [CrossRef] [PubMed]
- Dobson, A.J.; Purves, J.; Kamysz, W.; Rolff, J. Comparing Selection on S. Aureus between Antimicrobial Peptides and Common Antibiotics. PLoS ONE 2013, 8, e76521. [Google Scholar] [CrossRef] [PubMed]
- Lofton, H.; Pränting, M.; Thulin, E.; Andersson, D.I. Mechanisms and Fitness Costs of Resistance to Antimicrobial Peptides LL-37, CNY100HL and Wheat Germ Histones. PLoS ONE 2013, 8, e68875. [Google Scholar] [CrossRef]


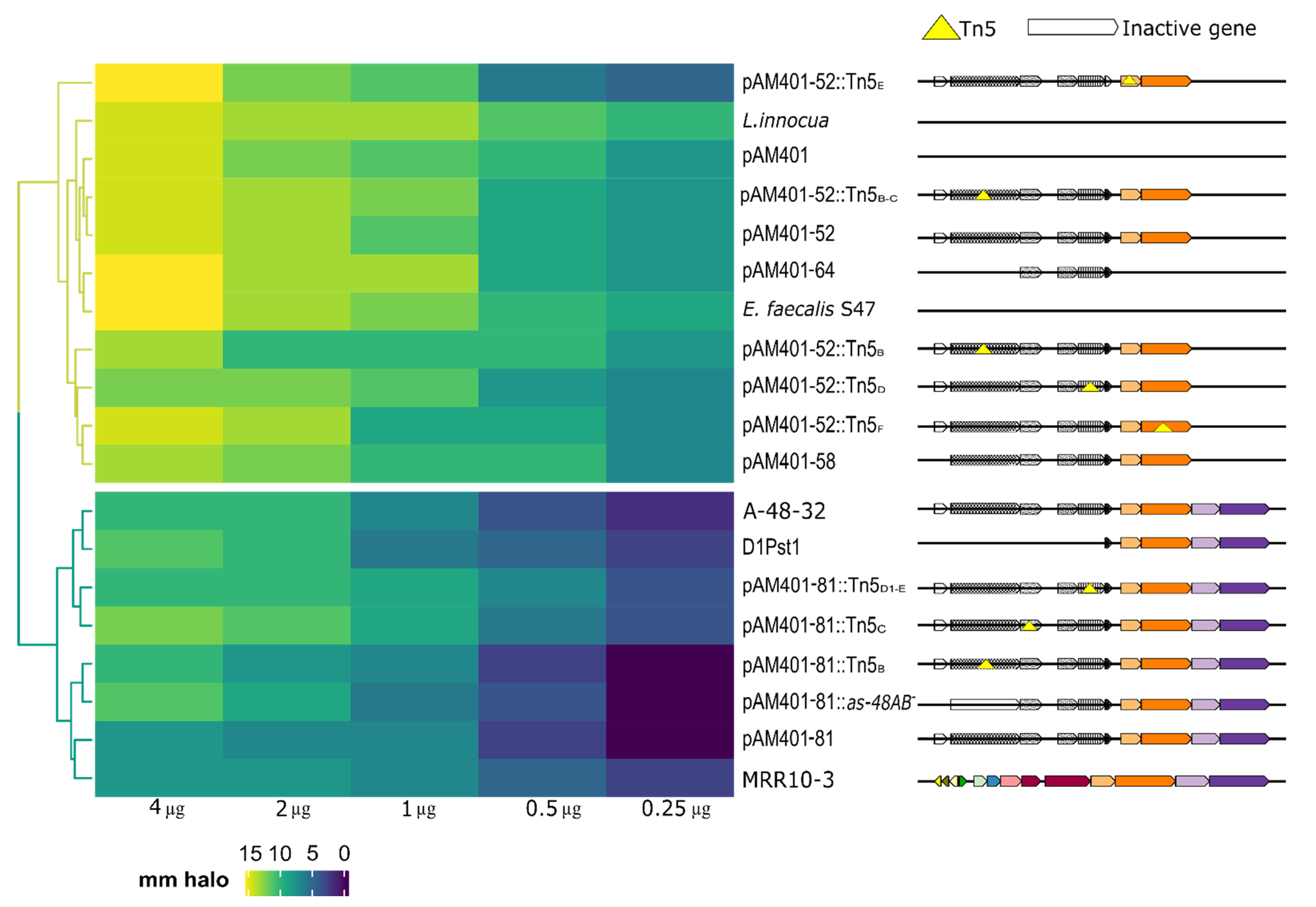
| Strain ** | Mutation | Reference or Source a |
|---|---|---|
| JH2-2 (pAM401-81) | as-48ABCC1DD1EFGH gene cluster cloned in pAM401 vector | [21] |
| D1Pst1 (as-48DEFGH cloned from 401-81::Tn5D1 mutant) | as-48D1EFGH | This study |
| JH2-2 (pAM401-81::Tn5D1-E) | as-48ABCC1DD1*EFGH | [21] |
| JH2-2 (pAM401-81::Tn5C) | as-48ABC*C1DD1EFGH | [35] |
| JH2-2 (pAM401-81::Tn5B) | as-48AB*CC1DD1EFGH | [35] |
| JH2-2 (pAM401-81::as-48AB-) | as-48CC1DD1EFGH | [21] |
| JH2-2 (pAM401-52) | as-48ABCC1DD1EF | [20] |
| JH2-2 (pAM401-52::Tn5F) | as-48ABCC1DD1EF* | [20] |
| JH2-2 (pAM401-52:: Tn5E) | as-48ABCC1DD1E*F | [20] |
| JH2-2 (pAM401-52::Tn5D) | as-48ABCC1D*D1EF | [20] |
| JH2-2 (pAM401-52::Tn5B-C) | as-48AB*CC1DD1EF | [20] |
| JH2-2 (pAM401-52::Tn5B) | as-48AB*CC1DD1EF | [20] |
| pAM401-58 (pAM401-52::as48A-) | as-48BCC1DD1EF | [35] |
| pAM401-64 | ABC transporter cloned into pAM401 as-48CC1DD1 | [20] |
| E. faecalis MRR10-3 | Wild type | [28] |
| E. faecium F58 | Wild type | [36] |
| E. faecalis A-48-32 | Wild type AS-48 producer | [18] |
| JH2-2 (pAM401) | Negative control | [37] |
| L.innocua 4030 | Wild type Indicator strain | CECT |
| E.faecalis S-47 | Wild type Indicator strain | [38] |
| As-48EFGH | Percentage Identity (%) | MR10EFGH |
|---|---|---|
| As-48E | 46.01 | Mr10E |
| As-48F | 97.74 | Mr10F |
| As-48G | 99.56 | Mr10G |
| As-48H | 95.55 | Mr10H |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Teso-Pérez, C.; Martínez-Bueno, M.; Peralta-Sánchez, J.M.; Valdivia, E.; Maqueda, M.; Fárez-Vidal, M.E.; Martín-Platero, A.M. Enterocin Cross-Resistance Mediated by ABC Transport Systems. Microorganisms 2021, 9, 1411. https://doi.org/10.3390/microorganisms9071411
Teso-Pérez C, Martínez-Bueno M, Peralta-Sánchez JM, Valdivia E, Maqueda M, Fárez-Vidal ME, Martín-Platero AM. Enterocin Cross-Resistance Mediated by ABC Transport Systems. Microorganisms. 2021; 9(7):1411. https://doi.org/10.3390/microorganisms9071411
Chicago/Turabian StyleTeso-Pérez, Claudia, Manuel Martínez-Bueno, Juan Manuel Peralta-Sánchez, Eva Valdivia, Mercedes Maqueda, M. Esther Fárez-Vidal, and Antonio M. Martín-Platero. 2021. "Enterocin Cross-Resistance Mediated by ABC Transport Systems" Microorganisms 9, no. 7: 1411. https://doi.org/10.3390/microorganisms9071411
APA StyleTeso-Pérez, C., Martínez-Bueno, M., Peralta-Sánchez, J. M., Valdivia, E., Maqueda, M., Fárez-Vidal, M. E., & Martín-Platero, A. M. (2021). Enterocin Cross-Resistance Mediated by ABC Transport Systems. Microorganisms, 9(7), 1411. https://doi.org/10.3390/microorganisms9071411






