Metagenomic Insight into the Community Structure of Maize-Rhizosphere Bacteria as Predicted by Different Environmental Factors and Their Functioning within Plant Proximity
Abstract
1. Introduction
2. Materials and Methods
2.1. Site Description and Soil Sampling
2.2. Physicochemical Analysis of Maize Rhizosphere and Bulk Soils
2.3. DNA Extraction and Metagenome Sequencing
2.4. Downstream Analysis of Sequences
2.5. Statistical Analysis
3. Results
3.1. Physicochemical Properties of Maize Rhizosphere and Bulk Soils
3.2. Metagenome Dataset of the Sampling Sites
3.3. Distribution of Bacterial Community in the Sampling Sites
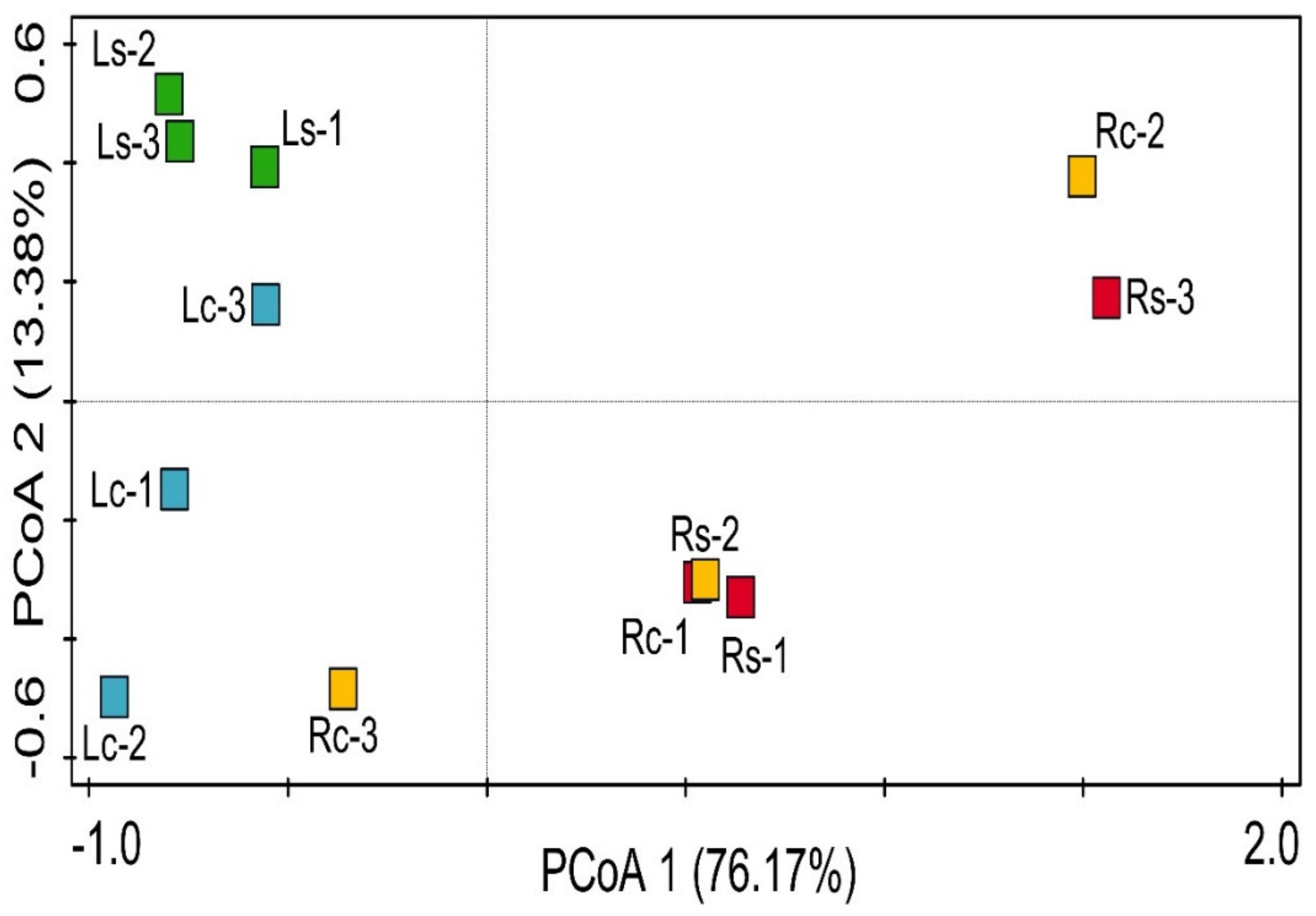
3.4. Structural Diversity of Rhizosphere Bacterial Communities across the Sampling Sites
3.5. Diversity Indices of Bacterial Communities across the Sampling Sites
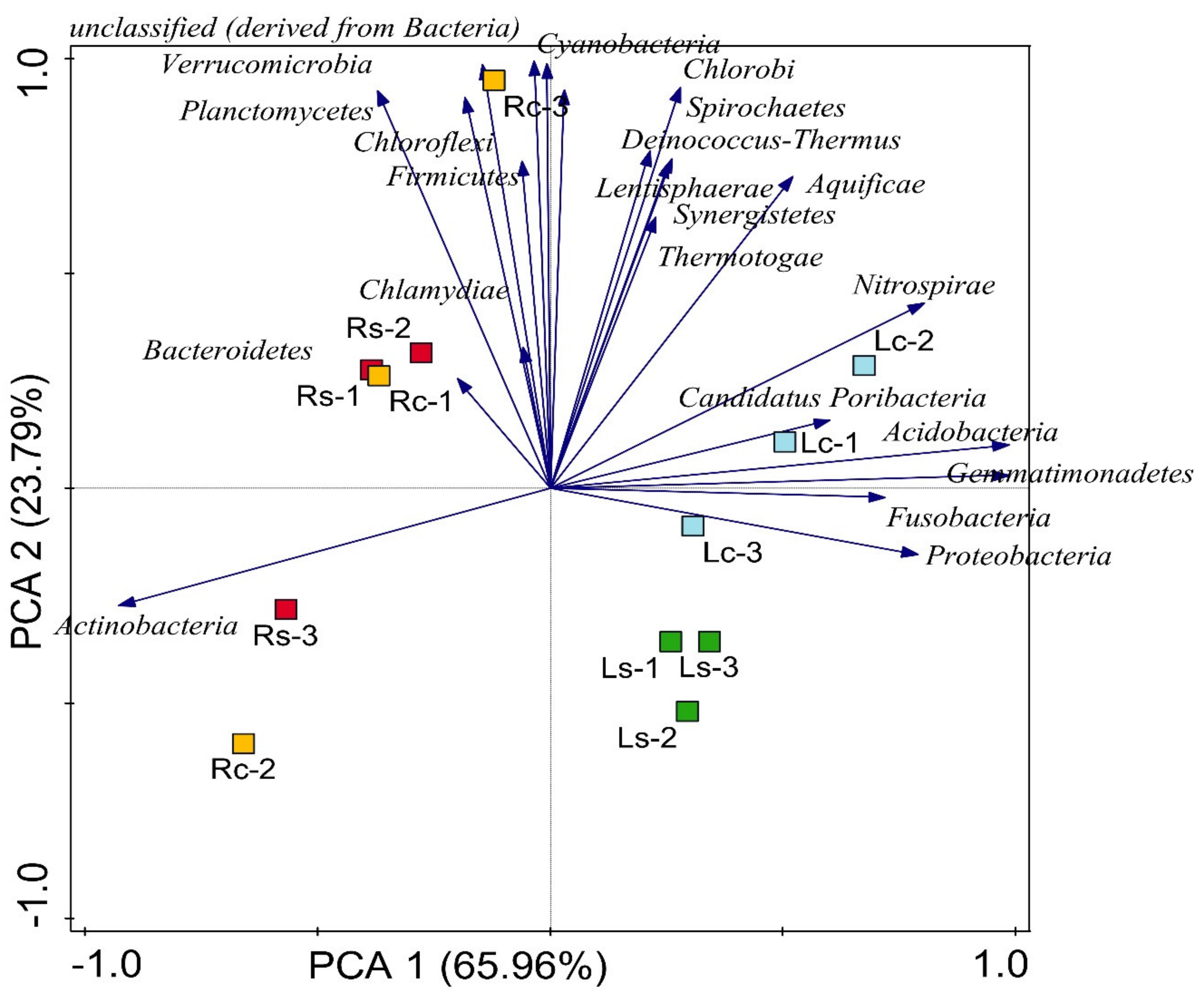
3.6. Influence of Environmental Variables on Bacterial Community
3.7. Metagenome-Based Functional Signatures of Bacteria Associated with the Maize Rhizosphere and Bulk Soils
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Beckers, B.; De Beeck, M.O.; Weyens, N.; Boerjan, W.; Vangronsveld, J. Structural variability and niche differentiation in the rhizosphere and endosphere bacterial microbiome of field-grown poplar trees. Microbiome 2017, 5, 25. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.A.; Kim, Y.; Kim, J.M.; Chu, B.; Joa, J.-H.; Sang, M.K.; Song, J.; Weon, H.-Y. A preliminary examination of bacterial, archaeal, and fungal communities inhabiting different rhizocompartments of tomato plants under real-world environments. J. Sci. Rep. 2019, 9, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Akinola, S.A.; Babalola, O.O. The importance of adverse soil microbiomes in the light of omics: Implications for food safety. Plant Soil Environ. 2020, 66, 421–430. [Google Scholar] [CrossRef]
- Coleman-Derr, D.; Desgarennes, D.; Fonseca-Garcia, C.; Gross, S.; Clingenpeel, S.; Woyke, T.; North, G.; Visel, A.; Partida-Martinez, L.P.; Tringe, S.G. Plant compartment and biogeography affect microbiome composition in cultivated and native Agave species. New Phytol. 2016, 209, 798–811. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Song, M.; Yang, L.; Zhang, D.; Sun, Y.; Shen, Z.; Luo, C.; Zhang, G. Exploring the influence of environmental factors on bacterial communities within the rhizosphere of the Cu-tolerant plant, Elsholtzia splendens. Sci. Rep. 2016, 6, 36302. [Google Scholar] [CrossRef]
- Compant, S.; Clément, C.; Sessitsch, A. Plant growth-promoting bacteria in the rhizo-and endosphere of plants: Their role, colonization, mechanisms involved and prospects for utilization. Soil Biol. Biochem. 2010, 42, 669–678. [Google Scholar] [CrossRef]
- Bulgarelli, D.; Garrido-Oter, R.; Münch, P.C.; Weiman, A.; Dröge, J.; Pan, Y.; McHardy, A.C.; Schulze-Lefert, P. Structure and function of the bacterial root microbiota in wild and domesticated barley. J. Cell Host Microbe 2015, 17, 392–403. [Google Scholar] [CrossRef]
- Buee, M.; De Boer, W.; Martin, F.; Van Overbeek, L.; Jurkevitch, E. The rhizosphere zoo: An overview of plant-associated communities of microorganisms, including phages, bacteria, archaea, and fungi, and of some of their structuring factors. Plant Soil 2009, 321, 189–212. [Google Scholar] [CrossRef]
- Verma, P.; Yadav, A.N.; Kumar, V.; Singh, D.P.; Saxena, A.K. Beneficial Plant-Microbes Interactions: Biodiversity of Microbes from Diverse Extreme Environments and its Impact for Crop Improvement Plant-Microbe Interactions in Agro-Ecological Perspectives; Springer: Cham, Switzerland, 2017; pp. 543–580. [Google Scholar]
- Fadiji, A.E.; Ayangbenro, A.S.; Babalola, O.O. Organic farming enhances the diversity and community structure of endophytic archaea and fungi in maize plant: A shotgun approach. J. Soil Sci. Plant Nutr. 2020, 20, 2587–2599. [Google Scholar] [CrossRef]
- Zhu, Y.-G.; Su, J.-Q.; Cao, Z.; Xue, K.; Quensen, J.; Guo, G.-X.; Yang, Y.-F.; Zhou, J.; Chu, H.-Y.; Tiedje, J.M. A buried Neolithic paddy soil reveals loss of microbial functional diversity after modern rice cultivation. Sci. Bull. 2016, 61, 1052–1060. [Google Scholar] [CrossRef]
- Pingali, P.L. Green revolution: Impacts, limits, and the path ahead. Proc. Nat. Acad. Sci. USA 2012, 109, 12302–12308. [Google Scholar] [CrossRef] [PubMed]
- Singh, H.; Reddy, M.S. Effect of inoculation with phosphate solubilizing fungus on growth and nutrient uptake of wheat and maize plants fertilized with rock phosphate in alkaline soils. Eur. J. Soil Biol. 2011, 47, 30–34. [Google Scholar] [CrossRef]
- Velivelli, S.L.; Sessitsch, A.; Prestwich, B.D. The role of microbial inoculants in integrated crop management systems. Potato Res. 2014, 57, 291–309. [Google Scholar] [CrossRef]
- Yarzábal, L.A.; Chica, E.J. Potential for developing low-input sustainable agriculture in the tropical Andes by making use of native microbial resources. In Plant-Microbe Interactions in Agro-Ecological Perspectives; Springer: Cham, Switzerland, 2017; pp. 29–54. [Google Scholar]
- Ghyselinck, J.; Velivelli, S.L.; Heylen, K.; O’Herlihy, E.; Franco, J.; Rojas, M.; De Vos, P.; Prestwich, B.D. Bioprospecting in potato fields in the Central Andean highlands: Screening of rhizobacteria for plant growth-promoting properties. System Appl. Microbiol. 2013, 36, 116–127. [Google Scholar] [CrossRef]
- Ke, X.; Feng, S.; Wang, J.; Lu, W.; Zhang, W.; Chen, M.; Lin, M. Effect of inoculation with nitrogen-fixing bacterium Pseudomonas stutzeri A1501 on maize plant growth and the microbiome indigenous to the rhizosphere. System Appl. Microbiol. 2019, 42, 248–260. [Google Scholar] [CrossRef]
- Tale, K.S.; Ingole, S. A review on role of physico-chemical properties in soil quality. J. Chem. Sci. Rev. Lett. 2015, 4, 57–66. [Google Scholar]
- Strickland, M.S.; Lauber, C.; Fierer, N.; Bradford, M.A. Testing the functional significance of microbial community composition. Ecology 2009, 90, 441–451. [Google Scholar] [CrossRef]
- Rousk, J.; Baath, E.; Brookes, P.C.; Lauber, C.L.; Lozupone, C.; Caporaso, J.G.; Knight, R.; Fierer, N. Soil bacterial and fungal communities across a pH gradient in an arable soil. ISME J. 2010, 4, 1340–1351. [Google Scholar] [CrossRef]
- Zhalnina, K.; Dias, R.; de Quadros, P.D.; Davis-Richardson, A.; Camargo, F.A.O.; Clark, I.M.; McGrath, S.P.; Hirsch, P.R.; Triplett, E.W. Soil pH determines microbial diversity and composition in the park grass experiment. Microb. Ecol. 2015, 69, 395–406. [Google Scholar] [CrossRef]
- Alori, E.T.; Babalola, O.O.; Prigent-Combaret, C. Impacts of microbial inoculants on the gowth and yield of maize plant. Open Agric. J. 2019, 13, 1–8. [Google Scholar] [CrossRef]
- International Maize and Wheat Improvement Center-CIMMYT. Maize in the World. CGIAR Research Prgram on Maize. 2016. Available online: https://maize.org/projects-cimmyt-and-iita-2/ (accessed on 15 November 2020).
- Du Plesis, J. Maize Production; ARC-Grain Crops Institute, Compiled by Directorate Agricultural Information Services; Department of Agriculture: Pretoria, South Africa, 2003; Available online: https://www.arc.agric.za/arc-gci/Pages/ARC-GCI-Homepage.aspx (accessed on 16 September 2020).
- Correa-Galeote, D.; Bedmar, E.J.; Fernández-González, A.J.; Fernández-López, M.; Arone, G.J. Bacterial communities in the rhizosphere of amilaceous maize (Zea mays L.) as assessed by pyrosequencing. Front. Plant Sci. 2016, 7, 1016. [Google Scholar] [CrossRef]
- Walters, W.A.; Jin, Z.; Youngblut, N.; Wallace, J.G.; Sutter, J.; Zhang, W.; González-Peña, A.; Peiffer, J.; Koren, O.; Shi, Q. Large-scale replicated field study of maize rhizosphere identifies heritable microbes. Proc. Nat. Acad. Sci. USA 2018, 115, 7368–7373. [Google Scholar] [CrossRef] [PubMed]
- De Araujo, A.S.F.; Miranda, A.R.L.; Sousa, R.S.; Mendes, L.W.; Antunes, J.E.L.; de Souza Oliveira, L.M.; de Araujo, F.F.; Melo, V.M.M.; Figueiredo, M.d.V.B. Bacterial community associated with rhizosphere of maize and cowpea in a subsequent cultivation. Appl. Soil Ecol. 2019, 143, 26–34. [Google Scholar] [CrossRef]
- Kudjordjie, E.N.; Sapkota, R.; Steffensen, S.K.; Fomsgaard, I.S.; Nicolaisen, M. Maize synthesized benzoxazinoids affect the host associated microbiome. Microbiome 2019, 7, 1–17. [Google Scholar] [CrossRef]
- South Africa Grain. The official Grain SA Magazine. 2020. Available online: http://sagrainmag.co.za/sa-grain-october-2020-e-book/ (accessed on 11 September 2020).
- Ranjan, R.; Rani, A.; Metwally, A.; McGee, H.S.; Perkins, D.L. Analysis of the microbiome: Advantages of whole genome shotgun versus 16S amplicon sequencing. Biochem. Biophys. Res. Commun. 2016, 469, 967–977. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Liu, Y.-X.; Zhang, N.; Hu, B.; Jin, T.; Xu, H.; Qin, Y.; Yan, P.; Zhang, X.; Guo, X. NRT1. 1B is associated with root microbiota composition and nitrogen use in field-grown rice. J. Nat. Biotechnol. 2019, 37, 676–684. [Google Scholar] [CrossRef]
- Walkley, A.; Black, I.A. An examination of the Degtjareff method for determining soil organic matter, and a proposed modification of the chromic acid titration method. J. Soil Sci. 1934, 37, 29–38. [Google Scholar] [CrossRef]
- Shi, J.-Y.; Yuan, X.-F.; Lin, H.-R.; Yang, Y.-Q.; Li, Z.-Y. Differences in soil properties and bacterial communities between the rhizosphere and bulk soil and among different production areas of the medicinal plant Fritillaria thunbergii. Int. J. Mol. Sci. 2011, 12, 3770–3785. [Google Scholar] [CrossRef] [PubMed]
- Deke, A.L.; Adugna, W.T.; Fite, A.T. Soil physico-chemical properties in termite mounds and adjacent control soil in Miyo and Yabello districts of Borana Zone, Southern Ethiopia. Am. J. Agric. Forest. 2016, 4, 69. [Google Scholar]
- Walker, D. Soil sulfate I. Extraction and measurement. Can. J. Soil Sci. 1972, 52, 253–260. [Google Scholar] [CrossRef]
- Pérez-Ruiz, C.L.; Badano, E.I.; Rodas-Ortiz, J.P.; Delgado-Sánchez, P.; Flores, J.; Douterlungne, D.; Flores-Cano, J.A. Climate change in forest ecosystems: A field experiment addressing the effects of raising temperature and reduced rainfall on early life cycle stages of oaks. Acta Oecol. 2018, 92, 35–43. [Google Scholar] [CrossRef]
- Baethgen, W.; Alley, M. A manual colorimetric procedure for measuring ammonium nitrogen in soil and plant Kjeldahl digests. Commun. Soil Sci. Plant Anal. 1989, 20, 961–969. [Google Scholar] [CrossRef]
- Meyer, F.; Paarmann, D.; D’Souza, M.; Olson, R.; Glass, E.M.; Kubal, M.; Paczian, T.; Rodriguez, A.; Stevens, R.; Wilke, A. The metagenomics RAST server–a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformat. 2008, 9, 1–8. [Google Scholar] [CrossRef]
- Kent, W.J. BLAT—the BLAST-like alignment tool. J. Gen. Res. 2002, 12, 656–664. [Google Scholar] [CrossRef]
- Wilke, A.; Harrison, T.; Wilkening, J.; Field, D.; Glass, E.M.; Kyrpides, N.; Mavrommatis, K.; Meyer, F. The M5nr: A novel non-redundant database containing protein sequences and annotations from multiple sources and associated tools. BMC Bioinformat. 2012, 13, 141. [Google Scholar] [CrossRef]
- Ondreičková, K.; Mihálik, D.; Ficek, A.; Hudcovicová, M.; Kraic, J.; Drahovská, H. Impact of genetically modified maize on the genetic diversity of rhizosphere bacteria: A two-year study in Slovakia. Pol. J. Ecol. 2014, 62, 67–76. [Google Scholar] [CrossRef]
- Mashiane, A.R.; Adeleke, R.A.; Bezuidenhout, C.C.; Chirima, G.J. Community composition and functions of endophytic bacteria of Bt maize. S. Afr. J. Sci. 2018, 114, 88–97. [Google Scholar] [CrossRef]
- Taffner, J.; Cernava, T.; Erlacher, A.; Berg, G. Novel insights into plant-associated archaea and their functioning in arugula (Eruca sativa Mill.). J. Adv. Res. 2019, 19, 39–48. [Google Scholar] [CrossRef] [PubMed]
- Enagbonma, B.J.; Babalola, O.O. Unveiling plant-beneficial function as seen in bacteria genes from termite mound soil. J. Soil Sci. Plant Nutr. 2020, 20, 421–430. [Google Scholar] [CrossRef]
- Yousuf, B.; Keshri, J.; Mishra, A.; Jha, B. Application of targeted metagenomics to explore abundance and diversity of CO2-fixing bacterial community using cbbL gene from the rhizosphere of Arachis hypogaea. Gene 2012, 506, 18–24. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Zheng, Y.; Ding, C.; Ren, X.; Yuan, J.; Sun, F.; Li, Y. Integrated metagenomics and molecular ecological network analysis of bacterial community composition during the phytoremediation of cadmium-contaminated soils by bioenergy crops. Ecotoxicol. Environ. Safety 2017, 145, 111–118. [Google Scholar] [CrossRef]
- Chica, E.; Buela, L.; Valdez, A.; Villena, P.; Peña, D.; Yarzábal, L.A. Metagenomic survey of the bacterial communities in the rhizosphere of three Andean tuber crops. Symbiosis 2019, 79, 141–150. [Google Scholar] [CrossRef]
- Berlanas, C.; Berbegal, M.; Elena, G.; Laidani, M.; Cibriain, J.F.; Sagües, A.; Gramaje, D. The fungal and bacterial rhizosphere microbiome associated with grapevine rootstock genotypes in mature and young vineyards. Front. Microbiol. 2019, 10, 1142. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Shao, C.; Chen, L.; Jin, X.; Ni, H. Plant growth-promoting effect of the chitosanolytic phosphate-solubilizing bacterium Burkholderia gladioli MEL01 after fermentation with chitosan and fertilization with rock phosphate. J. Plant Growth Regulat. 2020, 1–13. [Google Scholar] [CrossRef]
- Bhosale, P.; Bhagat, A.; Shete, B. Performance of different Mesorhizobium strains on nodulation, grain, straw yield and soil fertility status of chickpea. Int. J. Chem. Stud. 2020, 8, 2291–2294. [Google Scholar] [CrossRef]
- Daraz, U.; Li, Y.; Sun, Q.; Zhang, M.; Ahmad, I. Inoculation of Bacillus spp. modulate the soil bacterial communities and available nutrients in the rhizosphere of vetiver plant irrigated with acid mine drainage. Chemosphere 2020, 263, 128345. [Google Scholar]
- Wang, M.; Bian, Z.; Shi, J.; Wu, Y.; Yu, X.; Yang, Y.; Ni, H.; Chen, H.; Bian, X.; Li, T. Effect of the nitrogen-fixing bacterium Pseudomonas protegens CHA0-ΔretS-nif on garlic growth under different field conditions. Ind. Crop. Prod. 2020, 145, 111982. [Google Scholar] [CrossRef]
- Garrido-Sanz, D.; Redondo-Nieto, M.; Guirado, M.; Pindado, J.O.; Millán, R.; Martin, M.; Rivilla, R. Metagenomic insights into the bacterial functions of a diesel-degrading consortium for the rhizoremediation of diesel-polluted soil. Genes 2019, 10, 456. [Google Scholar] [CrossRef]
- Kumar, A.; Bisht, B.S.; Joshi, V.D. Biosorption of heavy metals by four acclimated microbial species, Bacillus spp., Pseudomonas spp., Staphylococcus spp., and Aspergillus niger. J. Biol. Environ. Sci. 2010, 4, 97–108. [Google Scholar]
- Ghazouani, S.; Béjaoui, Z.; Spiers, G.; Beckett, P.; Gtari, M.; Nkongolo, K. Effects of rhizobioaugmentation with N-Fixing Actinobacteria Frankia on metal mobility in Casuarina glauca-soil system irrigated with industrial wastewater: High level of metal exclusion of C. glauca. Water Air Soil Pollut. 2020, 231, 1–17. [Google Scholar] [CrossRef]
- Malisorn, K.; Chanchampa, S.; Kanchanasin, P.; Tanasupawat, S. Identification and plant growth-promoting activities of Proteobacteria isolated from root nodules and rhizospheric soils. Curr. Appl. Sci. Technol. 2020, 20, 479–493. [Google Scholar]
- Seballos, R.C.; Wyatt, K.H.; Bernot, R.J.; Brown, S.P.; Chandra, S.; Rober, A.R. Nutrient availability and organic matter quality shape bacterial community structure in a lake biofilm. Aquat. Microb. Ecol. 2020, 85, 1–18. [Google Scholar] [CrossRef]
- Berg, G.; Smalla, K. Plant species and soil type cooperatively shape the structure and function of microbial communities in the rhizosphere. FEMS Microbiol. Ecol. 2009, 68, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Schöps, R.; Goldmann, K.; Herz, K.; Lentendu, G.; Schöning, I.; Bruelheide, H.; Wubet, T.; Buscot, F. Land-use intensity rather than plant functional identity shapes bacterial and fungal rhizosphere communities. Front. Microbiol. 2018, 9, 2711. [Google Scholar] [CrossRef]
- Praeg, N.; Pauli, H.; Illmer, P. Microbial diversity in bulk and rhizosphere soil of Ranunculus glacialis along a high-alpine altitudinal gradient. Front. Microbiol. 2019, 10, 1429. [Google Scholar] [CrossRef]
- Arroyo, P.; de Miera, L.E.S.; Ansola, G. Influence of environmental variables on the structure and composition of soil bacterial communities in natural and constructed wetlands. J. Sci. Total Environ. 2015, 506, 380–390. [Google Scholar] [CrossRef] [PubMed]
- Enagbonma, B.J.; Ajilogba, C.F.; Babalola, O.O. Metagenomic profiling of bacterial diversity and community structure in termite mounds and surrounding soils. Arch. Microbiol. 2020, 202, 2697–2709. [Google Scholar] [CrossRef] [PubMed]
- Molefe, R.R.; Amoo, A.E.; Babalola, O.O. Metagenomic insights into the bacterial community structure and functional potentials in the rhizosphere soil of maize plants. J. Plant Interact. 2021, 16, 258–269. [Google Scholar] [CrossRef]
- Beatty, P.H.; Klein, M.S.; Fischer, J.J.; Lewis, I.A.; Muench, D.G.; Good, A.G. Understanding plant nitrogen metabolism through metabolomics and computational approaches. Plants 2016, 5, 39. [Google Scholar] [CrossRef] [PubMed]
- Baral, B.; Izaguirre-Mayoral, M.L. Purine-derived ureides under drought and salinity. Adv. Agron. 2017, 146, 167–204. [Google Scholar]
- Tajima, S.; Yatazawa, M.; Yamamoto, Y. Allantoin production and its utilization in relation to nodule formation in soybeans: Enzymatic studies. Soil Sci. Plant Nutr. 1977, 23, 225–235. [Google Scholar] [CrossRef]
- Pélissier, H.C.; Frerich, A.; Desimone, M.; Schumacher, K.; Tegeder, M. PvUPS1, an allantoin transporter in nodulated roots of French bean. Plant Physiol. 2004, 134, 664–675. [Google Scholar] [CrossRef] [PubMed]
- Lea, P.; Miflin, B. Photosynthetic Ammonia Assimilation Photosynthesis II; Springer: Berlin/Heidelberg, Germany, 1979; pp. 445–456. [Google Scholar]
- Lang, C.P.; Bárdos, G.; Merkt, N.; Zörb, C. Expression of key enzymes for nitrogen assimilation in grapevine rootstock in response to N-form and timing. J. Plant Nutr. Soil Sci. 2020, 183, 91–98. [Google Scholar] [CrossRef]
- Mendes, L.W.; Kuramae, E.E.; Navarrete, A.A.; Van Veen, J.A.; Tsai, S.M. Taxonomical and functional microbial community selection in soybean rhizosphere. ISME J. 2014, 8, 1577–1587. [Google Scholar] [CrossRef] [PubMed]
- Corpas, F.J.; Leterrier, M.; Valderrama, R.; Airaki, M.; Chaki, M.; Palma, J.M.; Barroso, J.B. Nitric oxide imbalance provokes a nitrosative response in plants under abiotic stress. Plant Sci. 2011, 181, 604–611. [Google Scholar] [CrossRef]
- Stigter, K.A.; Plaxton, W.C. Molecular mechanisms of phosphorus metabolism and transport during leaf senescence. Plants 2015, 4, 773–798. [Google Scholar] [CrossRef]
- Marschner, P.; Crowley, D.; Rengel, Z. Rhizosphere interactions between microorganisms and plants govern iron and phosphorus acquisition along the root axis–model and research methods. Soil Biol. Biochem. 2011, 43, 883–894. [Google Scholar] [CrossRef]
- Inoue, H.; Yamashita-Muraki, S.; Fujiwara, K.; Honda, K.; Ono, H.; Nonaka, T.; Kato, Y.; Matsuyama, T.; Sugano, S.; Suzuki, M. Fe2+ ions alleviate the symptom of citrus greening disease. Int. J. Mol. Sci. 2020, 21, 4033. [Google Scholar] [CrossRef]
- Ali, S.; Abbas, Z.; Seleiman, M.F.; Rizwan, M.; YavaŞ, İ.; Alhammad, B.A.; Shami, A.; Hasanuzzaman, M.; Kalderis, D. Glycine betaine accumulation, significance and interests for heavy metal tolerance in plants. Plants 2020, 9, 896. [Google Scholar] [CrossRef]
- Annunziata, M.G.; Ciarmiello, L.F.; Woodrow, P.; Dell’Aversana, E.; Carillo, P. Spatial and temporal profile of glycine betaine accumulation in plants under abiotic stresses. Front. Plant Sci. 2019, 10, 230. [Google Scholar] [CrossRef]
- Tang, D.; Wang, X.; Wang, J.; Wang, M.; Wang, Y.; Wang, W. Choline–betaine pathway contributes to hyperosmotic stress and subsequent lethal stress resistance in Pseudomonas protegens SN15-2. J. Biosci. 2020, 45, 1–10. [Google Scholar] [CrossRef]
- Boncompagni, E.; Østerås, M.; Poggi, M.-C.; le Rudulier, D. Occurrence of choline and glycine betaine uptake and metabolism in the family Rhizobiaceae and their roles in osmoprotection. Appl. Environ. Microbiol. 1999, 65, 2072–2077. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Ye, Z.; Zhao, J. RbrA, a cyanobacterial rubrerythrin, functions as a FNR-dependent peroxidase in heterocysts in protection of nitrogenase from damage by hydrogen peroxide in Anabaena sp. PCC 7120. Mol. Microbiol. 2007, 66, 1219–1230. [Google Scholar] [CrossRef] [PubMed]
- Rey, P.; Taupin-Broggini, M.; Couturier, J.; Vignols, F.; Rouhier, N. Is there a role for glutaredoxins and BOLAs in the perception of the cellular iron status in plants? Front. Plant Sci. 2019, 10, 712. [Google Scholar] [CrossRef] [PubMed]
- Rahman, M.M.; Asiri, A.M.; Khan, A. Importance of Selenium in the Environment and Human Health; IntechOpen: London, UK, 2020. [Google Scholar] [CrossRef]
- Rayman, M.P. Selenium and human health. Lancet 2012, 379, 1256–1268. [Google Scholar] [CrossRef]
- Di Gregorio, S.; Lampis, S.; Malorgio, F.; Petruzzelli, G.; Pezzarossa, B.; Vallini, G. Brassica juncea can improve selenite and selenate abatement in selenium contaminated soils through the aid of its rhizospheric bacterial population. Plant Soil 2006, 285, 233–244. [Google Scholar] [CrossRef]
- Silva, E.d.N.d.; Cidade, M.; Heerdt, G.; Ribessi, R.L.; Morgon, N.H.; Cadore, S. Effect of selenite and selenate application on mineral composition of lettuce plants cultivated under hydroponic conditions: Nutritional balance overview using a multifaceted study. J. Brazil. Chem. Soc. 2018, 29, 371–379. [Google Scholar] [CrossRef]
- Keto-Timonen, R.; Hietala, N.; Palonen, E.; Hakakorpi, A.; Lindström, M.; Korkeala, H. Cold shock proteins: A minireview with special emphasis on Csp-family of enteropathogenic Yersinia. Front. Microbiol. 2016, 7, 1151. [Google Scholar] [CrossRef]
- Shafqat, W.; Jaskani, M.J.; Maqbool, R.; Chattha, W.S.; Ali, Z.; Naqvi, S.A.; Haider, M.S.; Khan, I.A.; Vincent, C.I. Heat shock protein and aquaporin expression enhance water conserving behavior of citrus under water deficits and high temperature conditions. Environ. Exp. Bot. 2020, 181, 104270. [Google Scholar]
- Karlson, D.; Nakaminami, K.; Toyomasu, T.; Imai, R. A cold-regulated nucleic acid-binding protein of winter wheat shares a domain with bacterial cold shock proteins. J. Biol. Chem. 2002, 277, 35248–35256. [Google Scholar] [CrossRef]
- Sasaki, K.; Imai, R. Pleiotropic roles of cold shock domain proteins in plants. Front. Plant. Sci. 2012, 2, 116. [Google Scholar] [CrossRef] [PubMed]
- Reingruber, H.; Pontel, L.B. Formaldehyde metabolism and its impact on human health. Curr. Opin. Toxicol. 2018, 9, 28–34. [Google Scholar] [CrossRef]
- Martin, A.L. A comparison of the effects of tellurium and selenium on plants and animals. Am. J. Bot. 1937, 24, 198–203. [Google Scholar] [CrossRef]



| Sample Locations | Physicochemical Values | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| pH (H2O) | Sulfate (mg/kg) | Total C (%) | P (mg/kg) | K (mg/kg) | N-NO3 (mg/kg) | N-NH4 (mg/kg) | Org C (%) | OM (%) | |
| Ls | 5.62 ± 0.09 a | 1.60 ± 1.68 a | 0.90 ± 0.05 a | 50.98 ± 1.77 a | 240.00 ± 2.94 a | 16.29 ± 2.25 a | 3.61 ± 0.29 ab | 0.61 ± 0.02 a | 3.40 ± 0.16 a |
| Lc | 5.87 ± 0.22 a | 0.44 ± 0.36 a | 0.90 ± 0.01 a | 65.86 ± 13.71 a | 243.00 ± 0.82 a | 16.24 ± 0.59 a | 2.42 ± 0.19 a | 0.60 ± 0.01 a | 3.25 ± 0.03 a |
| Rs | 6.76 ± 0.28 b | 2.56 ± 2.66 a | 1.34 ± 0.24 a | 257.14 ± 35.32 b | 167.00 ± 11.63 b | 8.52 ± 2.68 b | 2.91 ± 1.12 a | 1.09 ± 0.09 b | 3.43 ± 0.39 a |
| Rc | 6.73 ± 0.26 b | 2.32 ± 2.75 a | 0.85 ± 0.50 a | 206.54 ± 81.73 b | 148.50 ± 34.95 b | 7.38 ± 2.46 b | 4.75 ± 1.21 b | 0.87 ± 0.15 c | 2.95 ± 0.85 a |
| p-values | ˂0.000 | 0.623 | 0.187 | 0.001 | ˂0.000 | 0.001 | 0.044 | ˂0.000 | 0.609 |
| Level | Indices | Ls | Rs | Lc | Rc | p-Value |
|---|---|---|---|---|---|---|
| Bacterial phylum | ||||||
| Shannon_H | 1.518 | 1.520 | 1.632 | 1.539 | 0.976 | |
| Evenness_e ˆ H/S | 0.207 | 0.208 | 0.233 | 0.212 | ||
| Class | ||||||
| Shannon_H | 1.964 | 1.895 | 2.051 | 1.919 | 0.949 | |
| Evenness_e ˆ H/S | 0.274 | 0.256 | 0.299 | 0.262 | ||
| Order | ||||||
| Shannon_H | 2.389 | 2.264 | 2.456 | 2.264 | 0.931 | |
| Evenness_e ˆ H/S | 0.363 | 0.321 | 0.389 | 0.321 | ||
| Family | ||||||
| Shannon_H | 3.179 | 3.268 | 3.200 | 3.325 | 0.544 | |
| Evenness_e ˆ H/S | 0.687 | 0.751 | 0.701 | 0.794 | ||
| Genus | ||||||
| Shannon_H | 3.109 | 3.155 | 3.064 | 3.188 | 0.860 | |
| Evenness_e ˆ H/S | 0.640 | 0.670 | 0.612 | 0.693 | ||
| Physicochemical Parameter | Contribution (%) | Pseudo-F | p-Value |
|---|---|---|---|
| N-NO3 | 82.70 | 9.50 | 0.019 |
| Sulfate | 9.70 | 1.30 | 0.50 |
| pH | 7.60 | ˂0.1 | 1.00 |
| SEED-Subsystem Level | Sampling Sites | |||||
|---|---|---|---|---|---|---|
| Level 2 | Level 3 | Ls | Rs | Lc | Rc | p-Value |
| Nitrogen Metabolism | Total Hits | 88,948 | 74,740 | 69,607 | 73,260 | |
| Relative Abundance (%) | ||||||
| Allantoin Utilization | 3.43 ± 0.10 a | 3.11 ± 0.10 b | 3.32 ± 0.06 ab | 3.13 ± 0.10 b | 0.02 | |
| Ammonia Assimilation | Ammonia Assimilation | 25.60 ± 0.37 a | 24.16 ± 0.27 b | 25.54 ± 0.27 a | 23.26 ± 0.41 b | 0.00 |
| Glutamate and Aspartate uptake in bacteria | 4.88 ± 0.33 a | 3.99 ± 0.32 b | 3.87 ± 0.27 ab | 4.12 ± 0.49 ab | 0.05 | |
| Glutamate dehydrogenase | 4.21 ± 0.08 a | 3.86 ± 0.51 a | 4.15 ± 0.08 a | 3.83 ± 0.24 a | 0.46 | |
| Glutamine, Glutamate, Aspartate and Asparagine Biosynthesis | 28.87 ± 0.14 a | 27.70 ± 0.30 a | 29.13 ± 0.10 a | 28.79 ± 1.57 a | 0.12 | |
| Glutamine Synthetases | 2.38 ± 0.03 ab | 2.28 ± 0.07 a | 2.49 ± 0.08 b | 2.50 ± 0.07 b | 0.02 | |
| Poly-gama-glutamate Biosynthesis | 2.65 ± 0.09 a | 2.19 ± 0.09 ab | 2.54 ± 0.12 a | 1.97 ± 0.35 b | 0.02 | |
| Cyanate Hydrolysis | 0.71 ± 0.01 a | 0.76 ± 0.14 a | 0.70 ± 0.04 a | 0.76 ± 0.08 a | 0.82 | |
| Denitrification | 1.95 ± 0.12 a | 2.72 ± 0.16 b | 5.40 ± 0.19 ab | 4.94 ± 0.35 ab | 0.01 | |
| Nitrate and Nitrite Ammonification | 15.86 ± 0.30 a | 20.98 ± 0.23 b | 14.97 ± 0.41 a | 18.00 ± 3.44 ab | 0.01 | |
| Nitric Oxide Synthase | 6.61 ± 0.13 a | 6.96 ± 0.43 a | 5.39 ± 0.02 b | 5.37 ± 0.30 b | 0.00 | |
| Nitrilase | 0.08 ± 0.01 a | 0.05 ± 0.02 a | 0.05 ± 0.01 a | 0.05 ± 0.01 a | 0.11 | |
| Nitrogen Fixation | 2.11 ± 0.07 ab | 2.35 ± 0.28 a | 1.52 ± 0.14 ab | 1.35 ± 0.52 b | 0.02 | |
| Nitrosative Stress | 0.77 ± 0.07 a | 0.87 ± 0.14 a | 1.85 ± 0.04 b | 1.93 ± 0.13 b | ˂0.00 | |
| Phosphorus Metabolism | Total Hits | 55,847 | 44,263 | 44,281 | 43,346 | |
| Relative Abundance (%) | ||||||
| Alkylphosphonate Utilization | 6.48 ± 0.13 a | 6.45 ± 0.53 a | 5.28 ± 0.23 b | 5.02 ± 0.29 b | 0.01 | |
| Alkylphosphonate (TC_3.A.1.9.1) | 1.55 ± 0.07 a | 1.81 ± 0.28 a | 1.49 ± 0.09 a | 1.68 ± 0.01 a | 0.22 | |
| High Affinity Phosphate Transporter and Control of PHO Regulon | 18.71 ± 0.12 a | 18.38 ± 0.15 a | 18.90 ± 0.35 a | 18.75 ± 0.50 a | 0.27 | |
| P uptake Cyanobacteria | 8.07 ± 0.09 a | 8.06 ± 0.13 a | 6.92 ± 0.17 b | 6.99 ± 0.37 b | 0.00 | |
| Phosphate-Binding DING Proteins | 0.39 ± 0.17 a | 0.19 ± 0.03 a | 0.14 ± 0.04 a | 0.17 ± 0.03 a | 0.10 | |
| Phosphate Metabolism | PhoR-PhoB two Component Regulatory | 2.50 ± 0.11 a | 2.47 ± 0.01 a | 3.55 ± 0.10 a | 3.32 ± 0.08 a | 0.12 |
| Phosphate Metabolism | 61.32 ± 0.23 a | 62.29 ± 1.12 a | 52.09 ± 0.38 b | 52.79 ± 1.04 b | ˂0.01 | |
| Phosphoenolpyruvate phosphomutase | 0.74 ± 0.08 a | 0.58 ± 0.17 a | 0.58 ± 0.13 a | 0.55 ± 0.21 a | 0.48 | |
| Phosphonate Metabolism | 1.25 ± 0.07 a | 0.77 ± 0.11 b | 1.05 ± 0.03 ab | 0.73 ± 0.37 ab | 0.04 | |
| Iron Acquisition and Metabolism | Total Hits | 26,796 | 21,718 | 18,688 | 20,450 | |
| Relative Abundance (%) | ||||||
| Siderophore | Siderophore_Achromobactin | 0.17 ± 0.14 a | 0.06 ± 0.01 a | 0.04 ± 0.03 a | 0.08 ± 0.02 a | 0.35 |
| Siderophore_Aerobactin | 0.09 ± 0.01 a | 0.18 ± 0.07 b | 0.06 ± 0.00 ab | 0.17 ± 0.03 ab | 0.05 | |
| Siderophore_Desferrioxamine_E | 0.38 ± 0.06 a | 0.76 ± 0.60 a | 0.27 ± 0.03 a | 0.53 ± 0.19 a | 0.48 | |
| Siderophore_Enterobactin | 0.28 ± 0.26 a | 0.16 ± 0.05 a | 0.11 ± 0.03 a | 0.19 ± 0.10 a | 0.68 | |
| Siderophore_Pyoverdine | 9.90 ± 0.73 a | 9.62 ± 1.05 a | 9.60 ± 0.10 a | 8.05 ± 2.08 a | 0.38 | |
| Siderophore_Staphylobactin | 0.06 ± 0.01 a | 0.02 ± 0.01 b | 0.07 ± 0.02 a | 0.01 ± 0.00 b | 0.00 | |
| Siderophore_Yersiniabactin_Biosynthesis | 1.27 ± 0.11 a | 1.53 ± 0.08 a | 1.41 ± 0.07 a | 1.19 ± 0.22 a | 0.08 | |
| Siderophore_Assembly_Kit | 5.96 ± 0.46 a | 8.21 ± 1.67 b | 5.89 ± 0.22 ab | 7.44 ± 0.25 ab | 0.05 | |
| Siderophore_Pyochelin | 0.22 ± 0.04 a | 0.28 ± 0.04 a | 0.19 ± 0.00 a | 0.25 ± 0.13 a | 0.47 | |
| Vibrioferrin_Synthesis | 0.17 ± 0.03 a | 0.12 ± 0.03 ab | 0.11 ± 0.02 ab | 0.05 ± 0.03 b | 0.02 | |
| Bacillibactin_Siderophore | 0.52 ± 0.05 a | 0.55 ± 0.03 a | 0.56 ± 0.03 a | 0.48 ± 0.08 a | 0.39 | |
| Transport of Iron | 9.71 ± 0.54 ab | 11.40 ± 0.70 a c | 9.01 ± 0.73 b | 12.30 ± 0.69 c | 0.01 | |
| ABC Type Iron Transport System | 3.26 ± 0.32 a | 1.88 ± 0.31 b | 3.19 ± 0.06 a | 1.86 ± 0.38 b | 0.00 | |
| ABC transporter Iron.B12.Siderophore.hemin | 0.71 ± 0.03 a | 0.98 ± 0.15 ab | 0.61 ± 0.10 b | 0.75 ± 0.16 ab | 0.05 | |
| Campylobacter Iron Metabolism | 12.16 ± 0.43 ab | 12.20 ± 0.70 a | 13.70 ± 0.29 b | 12.89 ± 0.06 ab | 0.04 | |
| Ferrous Iron Transporter EfeUOB Low pH Induced | 4.77 ± 0.41 a | 5.30 ± 1.37 a | 4.45 ± 0.33 a | 5.70 ± 2.54 a | 0.78 | |
| Hemen Hemin uptake and Utilization Systems (Gram Negative) | 8.93 ± 0.42 a | 7.67 ± 0.33 ab | 9.00 ± 0.45 b | 7.81 ± 0.91 ab | 0.05 | |
| Hemen Hemin uptake and Utilization Systems (Gram Positives) | 3.52 ± 0.39 a | 3.55 ± 0.29 a | 4.11 ± 0.45 a | 3.76 ± 0.09 a | 0.31 | |
| Hemin Transport System | 2.45 ± 0.21 a | 2.37 ± 0.34 a | 2.46 ± 0.06 a | 2.59 ± 0.18 a | 0.81 | |
| Iron III Dicitrate Transport System Fec | 0.94 ± 0.11 a | 1.10 ± 0.08 a | 0.81 ± 0.10 a | 1.10 ± 0.24 a | 0.15 | |
| Iron Scavenging Cluster in Thermus | 0.75 ± 0.07 a | 1.26 ± 0.27 b | 0.70 ± 0.00 a | 1.12 ± 0.12 ab | 0.02 | |
| Iron Acquisition in Streptococcus | 1.64 ± 0.06 a | 2.51 ± 0.21 b | 1.74 ± 0.22 a | 2.68 ± 0.07 b | 0.00 | |
| Iron Acquisition in Vibrio | 31.89 ± 1.48 a | 27.85 ± 2.06 ab | 31.66 ± 0.56 a | 28.64 ± 1.36 b | 0.05 | |
| Stress Response | Total Hits | 86,683 | 68,762 | 66,309 | 66,096 | |
| Relative Abundance (%) | ||||||
| SigmaB Stress Response Regulation | 6.36 ± 0.23 a | 6.40 ± 0.27 a | 6.59 ± 0.35 a | 6.33 ± 0.17 a | 0.74 | |
| Osmotic Stress | Synthesis of Osmoregulation Periplasmic glucans | 5.22 ± 0.13 a | 5.79 ± 1.30 a | 5.62 ± 0.07 a | 6.46 ± 1.88 a | 0.37 |
| Choline and Betaine Uptake and Biosynthesis | 8.97 ± 0.46 a | 11.03 ± 0.62 b | 8.42 ± 0.61 a | 9.85 ± 0.96 ab | 0.02 | |
| Ectoine Synthesis and Regulation | 1.23 ± 0.05 a | 0.33 ± 0.11 b | 0.22 ± 0.07 b | 0.26 ± 0.07 b | ˂0.00 | |
| Osmoprotection ABC Transporter YehZYXW of Enterobacteriales | 0.08 ± 0.04 a | 0.05 ± 0.01 a | 0.07 ± 0.00 a | 0.04 ± 0.01 a | 0.33 | |
| Osmoregulation | 1.70 ± 0.03 a | 2.02 ± 0.14 ab | 1.77 ± 0.01 ab | 2.08 ± 0.21 b | 0.03 | |
| Phage Shock Protein Operon | 1.54 ± 0.01 a | 1.18 ± 0.22 b | 1.46 ± 0.06 ab | 1.22 ± 0.08 ab | 0.05 | |
| Oxidative Stress | CoA-disulfide Reductase (EC_1.8.1.14) cluster | 1.28 ± 0.01 a | 1.33 ± 0.03 a | 0.26 ± 0.02 b | 0.30 ± 0.02 b | 0.04 |
| Rubrerythrin | 2.18 ± 0.03 a | 2.05 ± 0.02 a | 2.23 ± 0.03 a | 2.06 ± 0.28 a | 0.34 | |
| Glutaredoxins | 0.38 ± 0.02 a | 0.40 ± 0.03 a | 0.37 ± 0.00 a | 0.37 ± 0.02 a | 0.44 | |
| Glutathione: Biosynthesis and Gamma Glutamyl Cycle | 9.45 ± 0.32 a | 8.26 ± 0.35 ab | 9.82 ± 0.04 ab | 8.02 ± 1.31 ab | 0.04 | |
| Glutathione: non-redox Reaction | 4.76 ± 0.22 a | 4.30 ± 0.16 ab | 4.24 ± 0.24 ab | 4.03 ± 0.18 b | 0.03 | |
| Glutathione: redox Cycle | 1.84 ± 0.19 a | 1.84 ± 0.08 a | 1.79 ± 0.07 a | 1.81 ± 0.13 a | 0.97 | |
| Glutathione Analogs Mycothiol | 3.95 ± 0.12 a | 4.60 ± 0.51 a | 3.78 ± 0.31 a | 5.43 ± 2.33 a | 0.39 | |
| NADPH Quinone Oxidoreductase_2 | 0.38 ± 0.01 a | 0.31 ± 0.08 a | 0.36 ± 0.02 a | 0.38 ± 0.11 a | 0.57 | |
| Protection from Reactive Oxygen Species | 4.12 ± 0.10 a | 4.44 ± 0.31 a | 4.06 ± 0.11 a | 4.70 ± 0.57 a | 0.20 | |
| Regulation of Oxidative Stress Response | 10.21 ± 0.33 a | 9.75 ± 0.42 a | 10.25 ± 0.49 a | 9.31 ± 0.46 a | 0.15 | |
| Acid Stress | Acid Resistance Mechanisms | 2.12 ± 0.05 a | 2.58 ± 0.30 a | 2.41 ± 0.19 a | 2.53 ± 0.41 a | 0.23 |
| Glutamate Transporter involved in Acid tolerance in Streptococcus | 0.07 ± 0.02 a | 0.13 ± 0.01 a | 0.07 ± 0.01 a | 0.17 ± 0.08 a | 0.06 | |
| Detoxification (Toxin Stress) | Uptake of Selenate and Selenite | 3.85 ± 0.24 a | 3.54 ± 0.21 a | 3.32 ± 0.41 a | 3.73 ± 0.34 a | 0.34 |
| Glutathione-dependent Pathway of Formaldehyde Detoxification | 1.91 ± 0.09 a | 1.72 ± 0.13 ab | 1.80 ± 0.34 ab | 1.57 ± 0.63 b | 0.03 | |
| Tellurite Resistance Chromosomal Determinants | 0.03 ± 0.04 a | 0.01 ± 0.07 b | 0.01 ± 0.18 b | 0.02 ± 0.01 ab | 0.02 | |
| Cold Shock | Cold Shock CspA Family of Proteins | 3.11 ± 0.00 a | 2.61 ± 0.00 b | 2.12 ± 0.01 a c | 2.70 ± 0.01 b c | 0.01 |
| Heat Shock | Heat Shock dnaK Gene Cluster Extended | 20.49 ± 0.07 a | 20.37 ± 0.11 a | 21.40 ± 0.14 a | 20.74 ± 0.19 a | 0.08 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Akinola, S.A.; Ayangbenro, A.S.; Babalola, O.O. Metagenomic Insight into the Community Structure of Maize-Rhizosphere Bacteria as Predicted by Different Environmental Factors and Their Functioning within Plant Proximity. Microorganisms 2021, 9, 1419. https://doi.org/10.3390/microorganisms9071419
Akinola SA, Ayangbenro AS, Babalola OO. Metagenomic Insight into the Community Structure of Maize-Rhizosphere Bacteria as Predicted by Different Environmental Factors and Their Functioning within Plant Proximity. Microorganisms. 2021; 9(7):1419. https://doi.org/10.3390/microorganisms9071419
Chicago/Turabian StyleAkinola, Saheed Adekunle, Ayansina Segun Ayangbenro, and Olubukola Oluranti Babalola. 2021. "Metagenomic Insight into the Community Structure of Maize-Rhizosphere Bacteria as Predicted by Different Environmental Factors and Their Functioning within Plant Proximity" Microorganisms 9, no. 7: 1419. https://doi.org/10.3390/microorganisms9071419
APA StyleAkinola, S. A., Ayangbenro, A. S., & Babalola, O. O. (2021). Metagenomic Insight into the Community Structure of Maize-Rhizosphere Bacteria as Predicted by Different Environmental Factors and Their Functioning within Plant Proximity. Microorganisms, 9(7), 1419. https://doi.org/10.3390/microorganisms9071419







