In Vitro Anti-Biofilm Activity of Bacteriophage K (ATCC 19685-B1) and Daptomycin against Staphylococci
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains
2.1.1. Phenotypic Characterization
2.1.2. Genotypic Characterization
2.2. Susceptibility of Strains to Staphylococcus aureus subsp. aureus Bacteriophage K (ATCC® 19685B1TM)
2.2.1. Phage Propagation and Enumeration
2.2.2. Strains’ Susceptibility Testing to Bacteriophage K
2.3. Biofilm Formation Assay
2.4. Activity of Bacteriophage K (ATCC 19685-B1) and Daptomycin against Staphylococci
2.4.1. Crystal Violet Microtiter Plate Assay
2.4.2. Methylthiazoltetrazolium (MTT) Assay
2.4.3. Growth Curve (GC) Assay
2.5. Statistical Analysis
2.5.1. Activity of Bacteriophage K (ATCC 19685-B1) and Daptomycin against Staphylococci (CV and MTT Assays)
2.5.2. Growth Inhibitory Effect of Bacteriophage
3. Results
3.1. Phenotypes and Genotypes of Studied Strains
3.2. Activity of Bacteriophage K and Daptomycin
3.3. Growth Curves
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Zheng, Y.; He, L.; Asiamah, T.K.; Otto, M. Colonization of Medical Devices by Staphylococci. Environ. Microbiol. 2018, 20, 3141–3153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Otto, M. Staphylococcal Biofilms. Microbiol. Spectr. 2018, 6. [Google Scholar] [CrossRef]
- Tong, S.Y.C.; Davis, J.S.; Eichenberger, E.; Holland, T.L.; Fowler, V.G. Staphylococcus aureus Infections: Epidemiology, Pathophysiology, Clinical Manifestations, and Management. Clin. Microbiol. Rev. 2015, 28, 603–661. [Google Scholar] [CrossRef] [Green Version]
- Foster, T.J. Surface Proteins of Staphylococcus epidermidis. Front. Microbiol. 2020, 11, 1829. [Google Scholar] [CrossRef]
- Mack, D.; Fischer, W.; Krokotsch, A.; Leopold, K.; Hartmann, R.; Egge, H.; Laufs, R. The Intercellular Adhesin Involved in Biofilm Accumulation of Staphylococcus epidermidis Is a Linear Beta-1,6-Linked Glucosaminoglycan: Purification and Structural Analysis. J. Bacteriol. 1996, 178, 175–183. [Google Scholar] [CrossRef] [Green Version]
- Donlan, R.M.; Costerton, J.W. Biofilms: Survival Mechanisms of Clinically Relevant Microorganisms. Clin. Microbiol. Rev. 2002, 15, 167–193. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, A.S.; de Lencastre, H.; Garau, J.; Kluytmans, J.; Malhotra-Kumar, S.; Peschel, A.; Harbarth, S. Methicillin-Resistant Staphylococcus aureus. Nat. Rev. Dis. Primer 2018, 4, 18033. [Google Scholar] [CrossRef]
- McCarthy, H.; Rudkin, J.K.; Black, N.S.; Gallagher, L.; O’Neill, E.; O’Gara, J.P. Methicillin Resistance and the Biofilm Phenotype in Staphylococcus aureus. Front. Cell. Infect. Microbiol. 2015, 5, 1. [Google Scholar] [CrossRef] [Green Version]
- Verderosa, A.D.; Totsika, M.; Fairfull-Smith, K.E. Bacterial Biofilm Eradication Agents: A Current Review. Front. Chem. 2019, 7, 824. [Google Scholar] [CrossRef] [Green Version]
- Roux, S.; Valour, F.; Karsenty, J.; Gagnieu, M.-C.; Perpoint, T.; Lustig, S.; Ader, F.; Martha, B.; Laurent, F.; Chidiac, C.; et al. Daptomycin > 6 Mg/Kg/Day as Salvage Therapy in Patients with Complex Bone and Joint Infection: Cohort Study in a Regional Reference Center. BMC Infect. Dis. 2016, 16, 83. [Google Scholar] [CrossRef] [Green Version]
- Lambert, M. IDSA Guidelines on the Treatment of MRSA Infections in Adults and Children. Am. Fam. Physician 2011, 84, 455–463. [Google Scholar]
- Nakonieczna, A.; Cooper, C.J.; Gryko, R. Bacteriophages and Bacteriophage-Derived Endolysins as Potential Therapeutics to Combat Gram-Positive Spore Forming Bacteria. J. Appl. Microbiol. 2015, 119, 620–631. [Google Scholar] [CrossRef]
- Alves, D.R.; Gaudion, A.; Bean, J.E.; Perez Esteban, P.; Arnot, T.C.; Harper, D.R.; Kot, W.; Hansen, L.H.; Enright, M.C.; Jenkins, A.T.A. Combined Use of Bacteriophage K and a Novel Bacteriophage to Reduce Staphylococcus aureus Biofilm Formation. Appl. Environ. Microbiol. 2014, 80, 6694–6703. [Google Scholar] [CrossRef] [Green Version]
- Kelly, D.; McAuliffe, O.; Ross, R.P.; Coffey, A. Prevention of Staphylococcus aureus Biofilm Formation and Reduction in Established Biofilm Density Using a Combination of Phage K and Modified Derivatives. Lett. Appl. Microbiol. 2012, 54, 286–291. [Google Scholar] [CrossRef]
- EUCAST. European Committee on Antimicrobial Susceptibility Testing. Breakpoint Tables for Interpretation of MICs and Zone Diameters, Version 10.0; 2020. Available online: www.eucast.org/clinical_breakpoints/ (accessed on 12 December 2020).
- Kontos, F.; Petinaki, E.; Spiliopoulou, I.; Maniati, M.; Maniatis, A.N. Evaluation of a Novel Method Based on PCR Restriction Fragment Length Polymorphism Analysis of the tuf Gene for the Identification of Staphylococcus Species. J. Microbiol. Methods 2003, 55, 465–469. [Google Scholar] [CrossRef]
- Murakami, K.; Minamide, W.; Wada, K.; Nakamura, E.; Teraoka, H.; Watanabe, S. Identification of Methicillin-Resistant Strains of Staphylococci by Polymerase Chain Reaction. J. Clin. Microbiol. 1991, 29, 2240–2244. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Petinaki, E.; Arvaniti, A.; Dimitracopoulos, G.; Spiliopoulou, I. Detection of MecA, MecR1 and MecI Genes among Clinical Isolates of Methicillin-Resistant Staphylococci by Combined Polymerase Chain Reactions. J. Antimicrob. Chemother. 2001, 47, 297–304. [Google Scholar] [CrossRef] [Green Version]
- Cafiso, V.; Bertuccio, T.; Santagati, M.; Campanile, F.; Amicosante, G.; Perilli, M.G.; Selan, L.; Artini, M.; Nicoletti, G.; Stefani, S. Presence of the Ica Operon in Clinical Isolates of Staphylococcus epidermidis and Its Role in Biofilm Production. Clin. Microbiol. Infect. Off. Publ. Eur. Soc. Clin. Microbiol. Infect. Dis. 2004, 10, 1081–1088. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roche, F.M.; Massey, R.; Peacock, S.J.; Day, N.P.J.; Visai, L.; Speziale, P.; Lam, A.; Pallen, M.; Foster, T.J.Y. Characterization of Novel LPXTG-Containing Proteins of Staphylococcus aureus Identified from Genome Sequences. Microbiology 2003, 149, 643–654. [Google Scholar] [CrossRef] [Green Version]
- Gomes, A.R.; Vinga, S.; Zavolan, M.; de Lencastre, H. Analysis of the Genetic Variability of Virulence-Related Loci in Epidemic Clones of Methicillin-Resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 2005, 49, 366–379. [Google Scholar] [CrossRef] [Green Version]
- Vandecasteele, S.J.; Peetermans, W.E.; R Merckx, R.; Rijnders, B.J.A.; Van Eldere, J. Reliability of the Ica, Aap and AtlE Genes in the Discrimination between Invasive, Colonizing and Contaminant Staphylococcus epidermidis Isolates in the Diagnosis of Catheter-Related Infections. Clin. Microbiol. Infect. Off. Publ. Eur. Soc. Clin. Microbiol. Infect. Dis. 2003, 9, 114–119. [Google Scholar] [CrossRef] [Green Version]
- Giormezis, N.; Kolonitsiou, F.; Foka, A.; Drougka, E.; Liakopoulos, A.; Makri, A.; Papanastasiou, A.D.; Vogiatzi, A.; Dimitriou, G.; Marangos, M.; et al. Coagulase-Negative Staphylococcal Bloodstream and Prosthetic-Device-Associated Infections: The Role of Biofilm Formation and Distribution of Adhesin and Toxin Genes. J. Med. Microbiol. 2014, 63, 1500–1508. [Google Scholar] [CrossRef] [PubMed]
- Kropinski, A.; Mazzocco, A. Enumeration of Bacteriophages by Double Agar Overlay Plaque Assay. In Bacteriophages; Methods and Protocols; Clokie, M.R.J., Kropinski, A., Eds.; Volume 1: Isolation, Characterization, and Interactions; Humana Press: New Delhi, India, 2009; pp. 69–76. [Google Scholar]
- Orlova, E.V. Bacteriophages and Their Structural Organisation. In Bacteriophages; Kurtboke, I., Ed.; InTech: Rijeka, Croatia, 2012. [Google Scholar]
- Mazzocco, A.; Wadell, T.E. Enumeration of Bacteriophages Using the Small Drop Plaque Assay System. In Bacteriophages; Methods and Protocols; Clokie, M.R.J., Kropinski, A., Eds.; Volume 1: Isolation, Characterization, and Interactions; Humana Press: New Delhi, India, 2009; pp. 81–85. [Google Scholar]
- Stepanović, S.; Vuković, D.; Hola, V.; Di Bonaventura, G.; Djukić, S.; Cirković, I.; Ruzicka, F. Quantification of Biofilm in Microtiter Plates: Overview of Testing Conditions and Practical Recommendations for Assessment of Biofilm Production by Staphylococci. APMIS Acta Pathol. Microbiol. Immunol. Scand. 2007, 115, 891–899. [Google Scholar] [CrossRef] [PubMed]
- Tótoli, E.G.; Salgado, H.R.N. Rapid Turbidimetric Assay to Determine the Potency of Daptomycin in Lyophilized Powder. Pharmaceutics 2015, 7, 106–121. [Google Scholar] [CrossRef] [PubMed]
- Feoktistova, M.; Geserick, P.; Leverkus, M. Crystal Violet Assay for Determining Viability of Cultured Cells. Cold Spring Harb. Protoc. 2016, 2016, pdb.prot087379. [Google Scholar] [CrossRef]
- Grein, F.; Müller, A.; Scherer, K.M.; Liu, X.; Ludwig, K.C.; Klöckner, A.; Strach, M.; Sahl, H.-G.; Kubitscheck, U.; Schneider, T. Ca2+-Daptomycin Targets Cell Wall Biosynthesis by Forming a Tripartite Complex with Undecaprenyl-Coupled Intermediates and Membrane Lipids. Nat. Commun. 2020, 11, 1455. [Google Scholar] [CrossRef] [Green Version]
- Mascio, C.T.M.; Alder, J.D.; Silverman, J.A. Bactericidal Action of Daptomycin against Stationary-Phase and Nondividing Staphylococcus aureus Cells. Antimicrob. Agents Chemother. 2007, 51, 4255–4260. [Google Scholar] [CrossRef] [Green Version]
- van Meerloo, J.; Kaspers, G.J.L.; Cloos, J. Cell Sensitivity Assays: The MTT Assay. Methods Mol. Biol. Clifton NJ 2011, 731, 237–245. [Google Scholar] [CrossRef]
- Grela, E.; Kozłowska, J.; Grabowiecka, A. Current Methodology of MTT assay in Bacteria-A Review. Acta Histochem. 2018, 120, 303–311. [Google Scholar] [CrossRef]
- Bartomeu Garcia, C.; Shi, D.; Webster, T.J. Tat-Functionalized Liposomes for the Treatment of Meningitis: An in Vitro Study. Int. J. Nanomed. 2017, 12, 3009–3021. [Google Scholar] [CrossRef] [Green Version]
- Horowitz, J.; Normand, M.D.; Corradini, M.G.; Peleg, M. Probabilistic Model of Microbial Cell Growth, Division, and Mortality. Appl. Environ. Microbiol. 2010, 76, 230–242. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McCann, M.T.; Gilmore, B.F.; Gorman, S.P. Staphylococcus Epidermidis Device-Related Infections: Pathogenesis and Clinical Management. J. Pharm. Pharmacol. 2008, 60, 1551–1571. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Widerström, M. Significance of Staphylococcus epidermidis in Health Care-Associated Infections, from Contaminant to Clinically Relevant Pathogen: This Is a Wake-Up Call! J. Clin. Microbiol. 2016, 54, 1679–1681. [Google Scholar] [CrossRef] [Green Version]
- Wertheim, H.F.L.; Melles, D.C.; Vos, M.C.; van Leeuwen, W.; van Belkum, A.; Verbrugh, H.A.; Nouwen, J.L. The Role of Nasal Carriage in Staphylococcus aureus Infections. Lancet Infect. Dis. 2005, 5, 751–762. [Google Scholar] [CrossRef]
- Coates, R.; Moran, J.; Horsburgh, M.J. Staphylococci: Colonizers and Pathogens of Human Skin. Future Microbiol. 2014, 9, 75–91. [Google Scholar] [CrossRef]
- Kranjec, C.; Morales Angeles, D.; Torrissen Mårli, M.; Fernández, L.; García, P.; Kjos, M.; Diep, D.B. Staphylococcal Biofilms: Challenges and Novel Therapeutic Perspectives. Antibiotics 2021, 10, 131. [Google Scholar] [CrossRef]
- Giormezis, N.; Papakonstantinou, K.; Kolonitsiou, F.; Drougka, E.; Foka, A.; Sarrou, S.; Anastassiou, E.D.; Petinaki, E.; Spiliopoulou, I.; Giormezis, N.; et al. Biofilm Synthesis and Its Relationship with Genetic Characteristics in Clinical Methicillin-Resistant Staphylococci. AIMS Bioeng. 2015, 2, 375–386. [Google Scholar] [CrossRef]
- Tristan, A.; Ying, L.; Bes, M.; Etienne, J.; Vandenesch, F.; Lina, G. Use of Multiplex PCR To Identify Staphylococcus aureus Adhesins Involved in Human Hematogenous Infections. J. Clin. Microbiol. 2003, 41, 4465–4467. [Google Scholar] [CrossRef] [Green Version]
- Qin, Z.; Ou, Y.; Yang, L.; Zhu, Y.; Tolker-Nielsen, T.; Molin, S.; Qu, D. Role of Autolysin-Mediated DNA Release in Biofilm Formation of Staphylococcus epidermidis. Microbiol. Read. Engl. 2007, 153, 2083–2092. [Google Scholar] [CrossRef] [Green Version]
- Bertsche, U.; Weidenmaier, C.; Kuehner, D.; Yang, S.-J.; Baur, S.; Wanner, S.; Francois, P.; Schrenzel, J.; Yeaman, M.R.; Bayer, A.S. Correlation of Daptomycin Resistance in a Clinical Staphylococcus aureus Strain with Increased Cell Wall Teichoic Acid Production and D-Alanylation. Antimicrob. Agents Chemother. 2011, 55, 3922–3928. [Google Scholar] [CrossRef] [Green Version]
- Mishra, N.N.; Bayer, A.S.; Moise, P.A.; Yeaman, M.R.; Sakoulas, G. Reduced Susceptibility to Host-Defense Cationic Peptides and Daptomycin Coemerge in Methicillin-Resistant Staphylococcus aureus from Daptomycin-Naive Bacteremic Patients. J. Infect. Dis. 2012, 206, 1160–1167. [Google Scholar] [CrossRef]
- Sader, H.S.; Farrell, D.J.; Flamm, R.K.; Jones, R.N. Daptomycin Activity Tested against 164457 Bacterial Isolates from Hospitalised Patients: Summary of 8 Years of a Worldwide Surveillance Programme (2005–2012). Int. J. Antimicrob. Agents 2014, 43, 465–469. [Google Scholar] [CrossRef]
- Raad, I.; Hanna, H.; Jiang, Y.; Dvorak, T.; Reitzel, R.; Chaiban, G.; Sherertz, R.; Hachem, R. Comparative Activities of Daptomycin, Linezolid, and Tigecycline against Catheter-Related Methicillin-Resistant Staphylococcus Bacteremic Isolates Embedded in Biofilm. Antimicrob. Agents Chemother. 2007, 51, 1656–1660. [Google Scholar] [CrossRef] [Green Version]
- Bauer, J.; Siala, W.; Tulkens, P.M.; Van Bambeke, F. A Combined Pharmacodynamic Quantitative and Qualitative Model Reveals the Potent Activity of Daptomycin and Delafloxacin against Staphylococcus aureus Biofilms. Antimicrob. Agents Chemother. 2013, 57, 2726–2737. [Google Scholar] [CrossRef] [Green Version]
- Kirby, A.E.; Garner, K.; Levin, B.R. The Relative Contributions of Physical Structure and Cell Density to the Antibiotic Susceptibility of Bacteria in Biofilms. Antimicrob. Agents Chemother. 2012, 56, 2967–2975. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parra-Ruiz, J.; Vidaillac, C.; Rose, W.E.; Rybak, M.J. Activities of High-Dose Daptomycin, Vancomycin, and Moxifloxacin Alone or in Combination with Clarithromycin or Rifampin in a Novel in Vitro Model of Staphylococcus aureus Biofilm. Antimicrob. Agents Chemother. 2010, 54, 4329–4334. [Google Scholar] [CrossRef] [Green Version]
- Cafiso, V.; Bertuccio, T.; Spina, D.; Purrello, S.; Stefani, S. Tigecycline Inhibition of a Mature Biofilm in Clinical Isolates of Staphylococcus aureus: Comparison with Other Drugs. FEMS Immunol. Med. Microbiol. 2010, 59, 466–469. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- LaPlante, K.L.; Woodmansee, S. Activities of Daptomycin and Vancomycin Alone and in Combination with Rifampin and Gentamicin against Biofilm-Forming Methicillin-Resistant Staphylococcus aureus Isolates in an Experimental Model of Endocarditis. Antimicrob. Agents Chemother. 2009, 53, 3880–3886. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Song, L. Applicability of the MTT Assay for Measuring Viability of Cyanobacteria and Algae, Specifically for Microcystis aeruginosa (Chroococcales, Cyanobacteria). Phycologia 2007, 46, 593–599. [Google Scholar] [CrossRef]
- Kraushaar, B.; Thanh, M.D.; Hammerl, J.A.; Reetz, J.; Fetsch, A.; Hertwig, S. Isolation and Characterization of Phages with Lytic Activity against Methicillin-Resistant Staphylococcus aureus Strains Belonging to Clonal Complex 398. Arch. Virol. 2013, 158, 2341–2350. [Google Scholar] [CrossRef] [PubMed]
- Capparelli, R.; Parlato, M.; Borriello, G.; Salvatore, P.; Iannelli, D. Experimental Phage Therapy against Staphylococcus aureus in Mice. Antimicrob. Agents Chemother. 2007, 51, 2765–2773. [Google Scholar] [CrossRef] [Green Version]
- Gupta, R.; Prasad, Y. Efficacy of Polyvalent Bacteriophage P-27/HP to Control Multidrug Resistant Staphylococcus aureus Associated with Human Infections. Curr. Microbiol. 2011, 62, 255–260. [Google Scholar] [CrossRef]
- Abedon, S.T. Bacteriophages and Biofilms: Ecology, Phage Therapy, Plaques; Nova Science: New York, NY, USA, 2011; ISBN 978-1-61761-588-7. [Google Scholar]
- Górski, A.; Miedzybrodzki, R.; Borysowski, J.; Weber-Dabrowska, B.; Lobocka, M.; Fortuna, W.; Letkiewicz, S.; Zimecki, M.; Filby, G. Bacteriophage Therapy for the Treatment of Infections. Curr. Opin. Investig. Drugs 2009, 10, 766–774. [Google Scholar]
- Lungren, M.P.; Christensen, D.; Kankotia, R.; Falk, I.; Paxton, B.E.; Kim, C.Y. Bacteriophage K for Reduction of Staphylococcus aureus biofilm on Central Venous Catheter Material. Bacteriophage 2013, 3, e26825. [Google Scholar] [CrossRef] [Green Version]
- Morrisette, T.; Lev, K.L.; Kebriaei, R.; Abdul-Mutakabbir, J.C.; Stamper, K.C.; Morales, S.; Lehman, S.M.; Canfield, G.S.; Duerkop, B.A.; Arias, C.A.; et al. Bacteriophage-Antibiotic Combinations for Enterococcus faecium with Varying Bacteriophage and Daptomycin Susceptibilities. Antimicrob. Agents Chemother. 2020, 64, e00993-20. [Google Scholar] [CrossRef]
- Duarte, A.C.; Fernández, L.; De Maesschalck, V.; Gutiérrez, D.; Campelo, A.B.; Briers, Y.; Lavigne, R.; Rodríguez, A.; García, P. Synergistic Action of Phage PhiIPLA-RODI and Lytic Protein CHAPSH3b: A Combination Strategy to Target Staphylococcus aureus Biofilms. NPJ Biofilms Microbiomes 2021, 7, 39. [Google Scholar] [CrossRef]

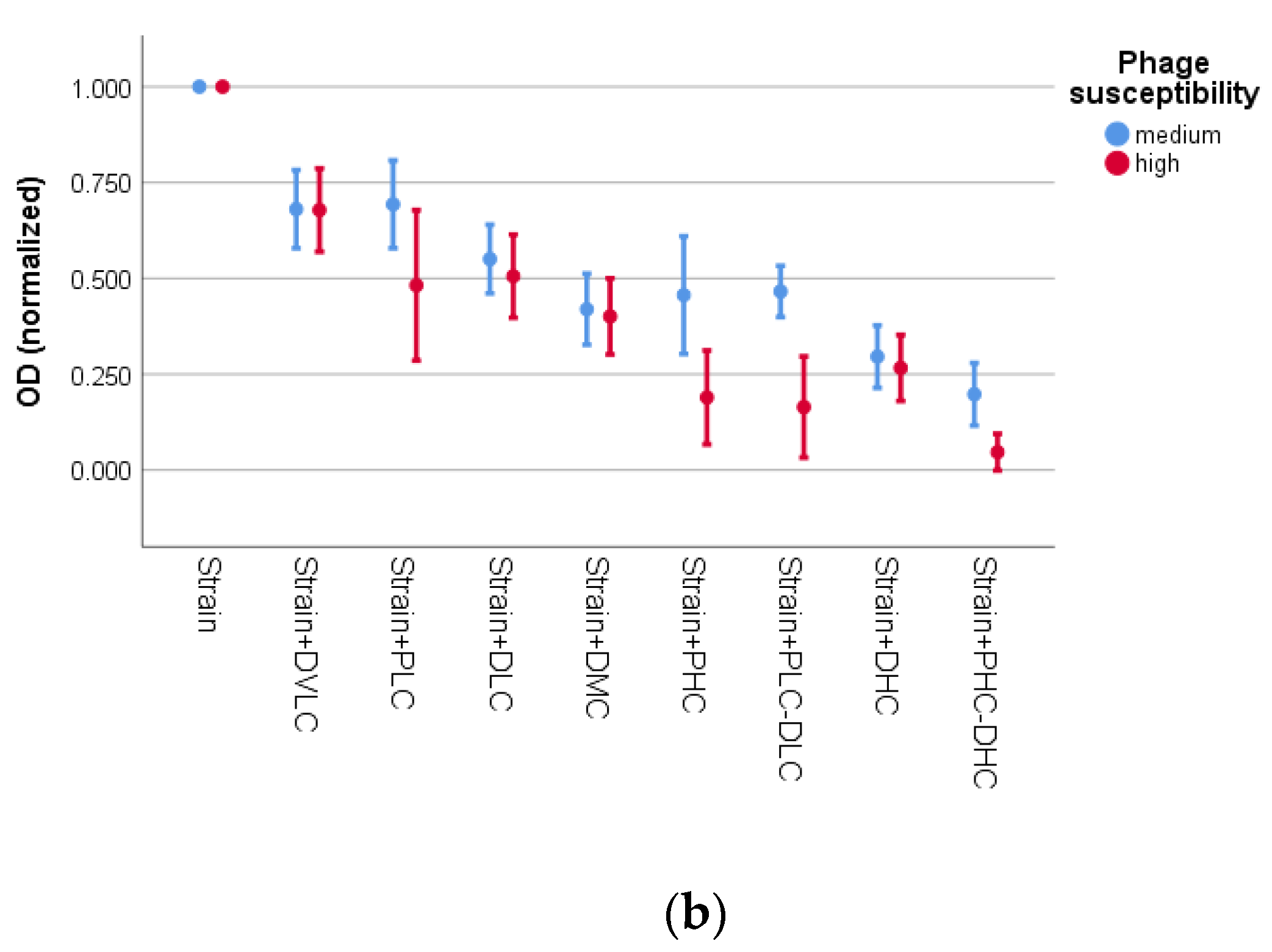
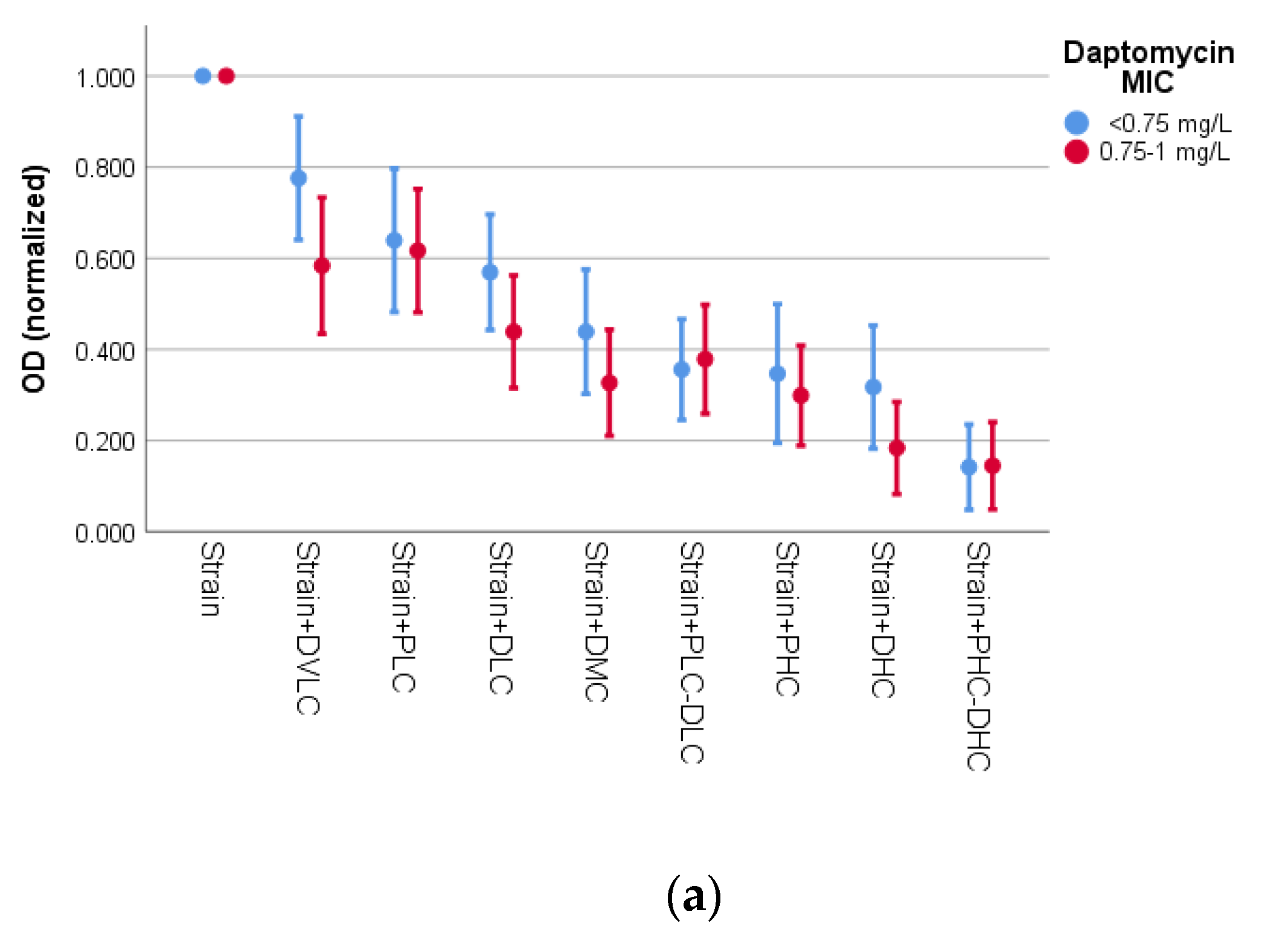

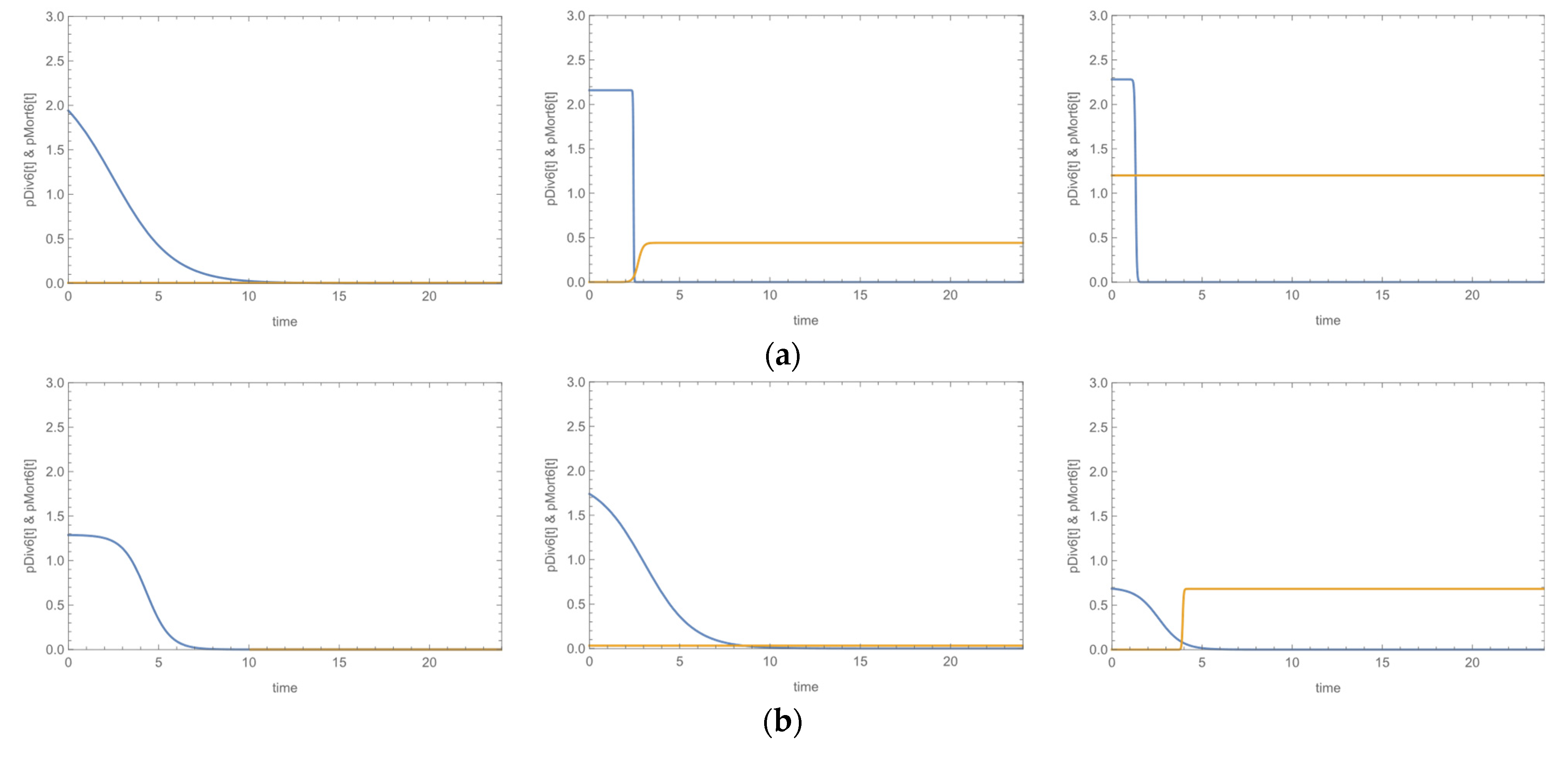
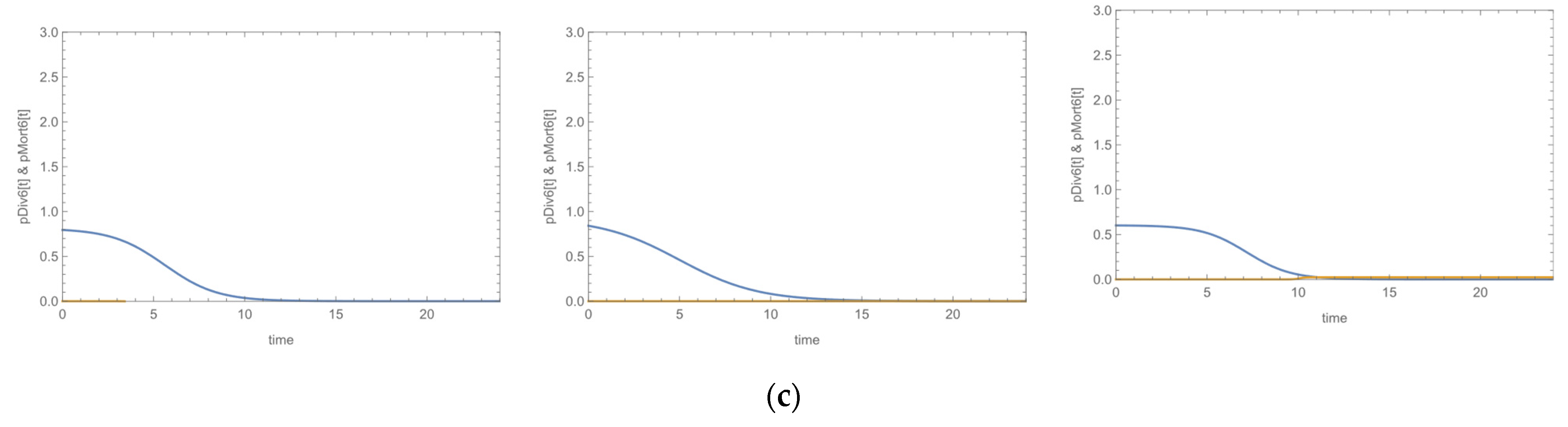
| Species | Biofilm Formation | MR | MDR | ica-Positive | fnbA-Positive | sasG-Positive | fbe-Positive | atlE-POSITIVE | Bacteriophage Susceptibility | Daptomycin Susceptibility | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Medium (80–100 pfu) | High (>100 pfu) | MIC < 0.75 mg/L | MIC 0.75–1 mg/L | |||||||||
| N | N | N | N | N | N | N | N | N | N | N | ||
| S. aureus (N = 10) | Positive (N = 8) | 1 | 1 | 4 | 8 | 6 | - | - | 3 | 5 | 5 | 3 |
| Negative (N = 2) | 1 | 0 | 0 | 1 | 1 | - | - | 1 | 1 | 2 | 0 | |
| S. epidermidis (N = 10) | Positive (N = 6) | 4 | 4 | 6 | - | - | 6 | 6 | 4 | 2 | 3 | 3 |
| Negative (N = 4) | 3 | 3 | 0 | - | - | 1 | 1 | 2 | 2 | 1 | 3 | |
| Total | 20 | 9 | 8 | 10 | 9 | 7 | 7 | 7 | 10 | 10 | 11 | 9 |
| Variable | O.R. 1 | 95 % CI 2 | p-Values | |
|---|---|---|---|---|
| Species | S. epidermidis | 1.0 | ||
| S. aureus | 4.44 | 0.69–28.59 | 0.117 | |
| Bacteriophage concentration | Low concentration (PLC) | 1.0 | ||
| High concentration (PHC) | 2.74 | 0.52–14.48 | 0.236 | |
| Biofilm formation | Positive | 1.0 | ||
| Negative | 7.52 | 0.71–79.26 | 0.093 | |
| Susceptibility to phage | 80–100 pfu | 1.0 | ||
| >100 pfu | 10.29 | 1.75–60.50 | 0.010 | |
| Daptomycin MIC | <0.75 mg/L | 1.0 | ||
| 0.75–1 mg/L | 0.81 | 0.11–6.03 | 0.840 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Plota, M.; Sazakli, E.; Giormezis, N.; Gkartziou, F.; Kolonitsiou, F.; Leotsinidis, M.; Antimisiaris, S.G.; Spiliopoulou, I. In Vitro Anti-Biofilm Activity of Bacteriophage K (ATCC 19685-B1) and Daptomycin against Staphylococci. Microorganisms 2021, 9, 1853. https://doi.org/10.3390/microorganisms9091853
Plota M, Sazakli E, Giormezis N, Gkartziou F, Kolonitsiou F, Leotsinidis M, Antimisiaris SG, Spiliopoulou I. In Vitro Anti-Biofilm Activity of Bacteriophage K (ATCC 19685-B1) and Daptomycin against Staphylococci. Microorganisms. 2021; 9(9):1853. https://doi.org/10.3390/microorganisms9091853
Chicago/Turabian StylePlota, Maria, Eleni Sazakli, Nikolaos Giormezis, Foteini Gkartziou, Fevronia Kolonitsiou, Michalis Leotsinidis, Sophia G. Antimisiaris, and Iris Spiliopoulou. 2021. "In Vitro Anti-Biofilm Activity of Bacteriophage K (ATCC 19685-B1) and Daptomycin against Staphylococci" Microorganisms 9, no. 9: 1853. https://doi.org/10.3390/microorganisms9091853
APA StylePlota, M., Sazakli, E., Giormezis, N., Gkartziou, F., Kolonitsiou, F., Leotsinidis, M., Antimisiaris, S. G., & Spiliopoulou, I. (2021). In Vitro Anti-Biofilm Activity of Bacteriophage K (ATCC 19685-B1) and Daptomycin against Staphylococci. Microorganisms, 9(9), 1853. https://doi.org/10.3390/microorganisms9091853








