Seeking a Role for Translational Control by Alternative Polyadenylation in Saccharomyces cerevisiae
Abstract
:1. Introduction
2. The Regulatory Features Impacted by APA
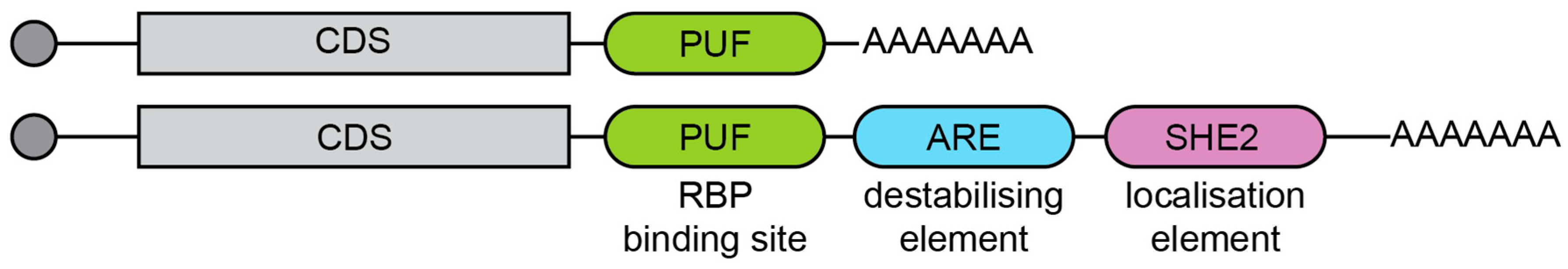
2.1. Cis-Regulatory Features
2.2. Trans-Regulatory Factors
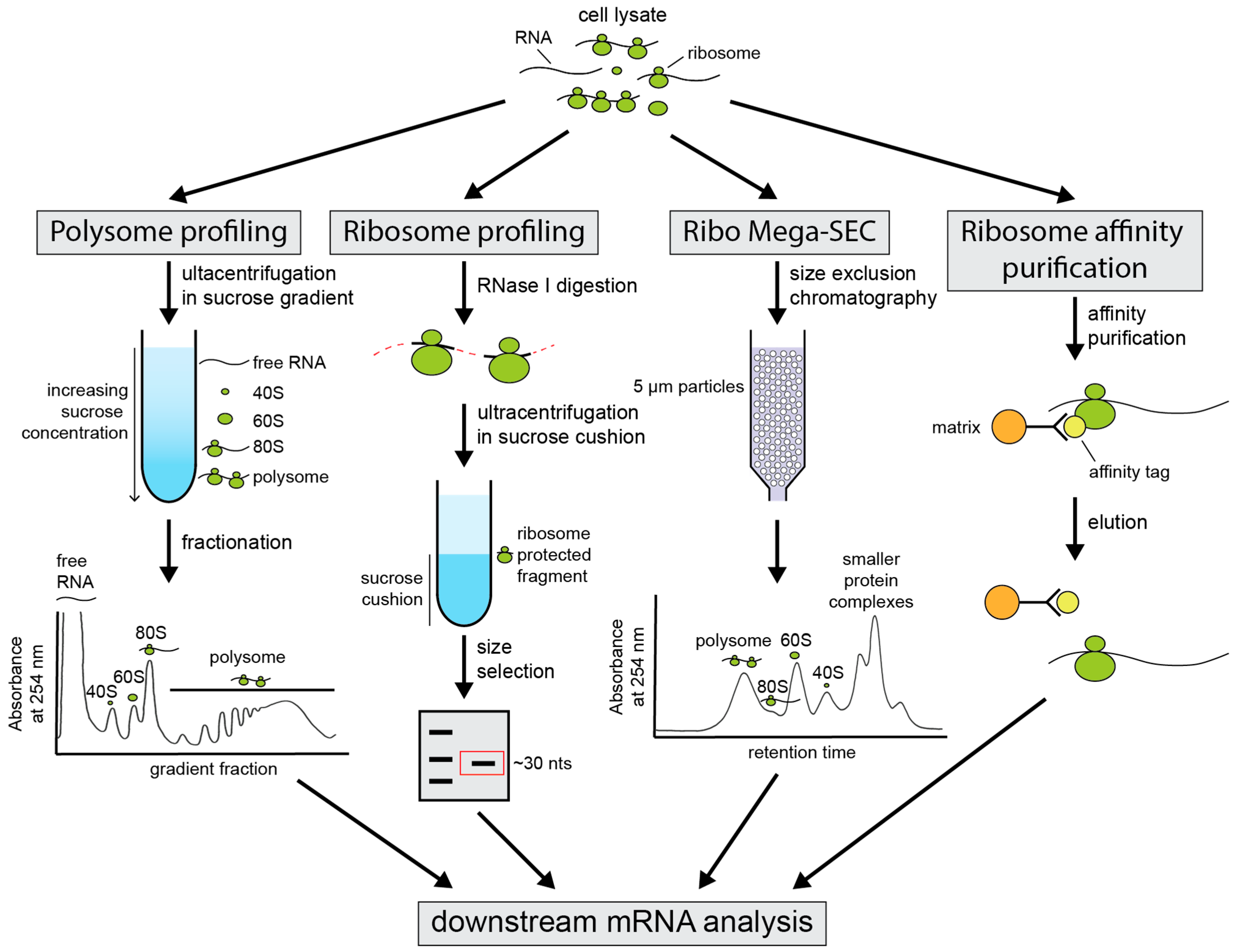
3. Techniques for Investigation of Alternative 3′ UTR Isoform Translation
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ozsolak, F.; Kapranov, P.; Foissac, S.; Kim, S.W.; Fishilevich, E.; Monaghan, A.P.; John, B.; Milos, P.M. Comprehensive Polyadenylation Site Maps in Yeast and Human Reveal Pervasive Alternative Polyadenylation. Cell 2010, 143, 1018–1029. [Google Scholar] [CrossRef] [Green Version]
- Turner, R.E.; Harrison, P.F.; Swaminathan, A.; Kraupner-Taylor, C.A.; Goldie, B.J.; See, M.; Peterson, A.L.; Schittenhelm, R.B.; Powell, D.R.; Creek, D.J.; et al. Genetic and pharmacological evidence for kinetic competition between alternative poly(A) sites in yeast. eLife 2021, 10, e65331. [Google Scholar] [CrossRef]
- Yague-Sanz, C.; Vanrobaeys, Y.; Fernandez, R.; Duval, M.; LaRochelle, M.; Beaudoin, J.; Berro, J.; Labbé, S.; Jacques, P.; Bachand, F. Nutrient-dependent control of RNA polymerase II elongation rate regulates specific gene expression programs by alternative polyadenylation. Genes Dev. 2020, 34, 883–897. [Google Scholar] [CrossRef] [PubMed]
- Geisberg, J.V.; Moqtaderi, Z.; Struhl, K. The transcriptional elongation rate regulates alternative polyadenylation in yeast. eLife 2020, 9, e59810. [Google Scholar] [CrossRef]
- Liu, X.; Hoque, M.; LaRochelle, M.; Lemay, J.-F.; Yurko, N.; Manley, J.L.; Bachand, F.; Tian, B. Comparative analysis of alternative polyadenylation in S. cerevisiae and S. pombe. Genome Res. 2017, 27, 1685–1695. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Michaels, K.K.; Mostafa, S.M.; Capella, J.R.; Moore, C.L. Regulation of alternative polyadenylation in the yeast Saccharomyces cerevisiae by histone H3K4 and H3K36 methyltransferases. Nucleic Acids Res. 2020, 48, 5407–5425. [Google Scholar] [CrossRef]
- Tian, B.; Manley, J. Alternative polyadenylation of mRNA precursors. Nat. Rev. Mol. Cell Biol. 2016, 18, 18–30. [Google Scholar] [CrossRef] [PubMed]
- Sandberg, R.; Neilson, J.R.; Sarma, A.; Sharp, P.A.; Burge, C.B. Proliferating Cells Express mRNAs with Shortened 3’ Untranslated Regions and Fewer MicroRNA Target Sites. Science 2008, 320, 1643–1647. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mayr, C.; Bartel, D.P. Widespread Shortening of 3′UTRs by Alternative Cleavage and Polyadenylation Activates Oncogenes in Cancer Cells. Cell 2009, 138, 673–684. [Google Scholar] [CrossRef] [Green Version]
- Lee, H.-C.; Li, L.; Gu, W.; Xue, Z.; Crosthwaite, S.K.; Pertsemlidis, A.; Lewis, Z.; Freitag, M.; Selker, E.U.; Mello, C.C.; et al. Diverse Pathways Generate MicroRNA-like RNAs and Dicer-Independent Small Interfering RNAs in Fungi. Mol. Cell 2010, 38, 803–814. [Google Scholar] [CrossRef] [Green Version]
- Drinnenberg, I.A.; Weinberg, D.E.; Xie, K.T.; Mower, J.P.; Wolfe, K.H.; Fink, G.R.; Bartel, D.P. RNAi in Budding Yeast. Science 2009, 326, 544–550. [Google Scholar] [CrossRef] [Green Version]
- Amrani, N.; Ghosh, S.; Mangus, D.A.; Jacobson, A. Translation factors promote the formation of two states of the closed-loop mRNP. Nature 2008, 453, 1276–1280. [Google Scholar] [CrossRef] [Green Version]
- Tanguay, R.L.; Gallie, D.R. Translational efficiency is regulated by the length of the 3’ untranslated region. Mol. Cell. Biol. 1996, 16, 146–156. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, M.; Sha, H.; Gao, Y.; Zeng, H.; Zhu, M.; Gao, X. Alternative 3′ UTR polyadenylation of Bzw1 transcripts display differential translation efficiency and tissue-specific expression. Biochem. Biophys. Res. Commun. 2006, 345, 479–485. [Google Scholar] [CrossRef]
- Di Giammartino, D.C.; Nishida, K.; Manley, J.L. Mechanisms and Consequences of Alternative Polyadenylation. Mol. Cell 2011, 43, 853–866. [Google Scholar] [CrossRef] [Green Version]
- Chen, T.; Van Steensel, B. Comprehensive analysis of nucleocytoplasmic dynamics of mRNA in Drosophila cells. PLoS Genet. 2017, 13, e1006929. [Google Scholar] [CrossRef] [Green Version]
- Bensidoun, P.; Zenklusen, D.; Oeffinger, M. Choosing the right exit: How functional plasticity of the nuclear pore drives selective and efficient mRNA export. Wiley Interdiscip. Rev. RNA 2021, e1660. [Google Scholar] [CrossRef]
- Sharova, L.V.; Sharov, A.A.; Nedorezov, T.; Piao, Y.; Shaik, N.; Ko, M.S. Database for mRNA Half-Life of 19 977 Genes Obtained by DNA Microarray Analysis of Pluripotent and Differentiating Mouse Embryonic Stem Cells. DNA Res. 2008, 16, 45–58. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, C.-Y.A.; Shyu, A.-B. AU-rich elements: Characterization and importance in mRNA degradation. Trends Biochem. Sci. 1995, 20, 465–470. [Google Scholar] [CrossRef]
- Vlasova, I.A.; Tahoe, N.M.; Fan, D.; Larsson, O.; Rattenbacher, B.; SternJohn, J.R.; Vasdewani, J.; Karypis, G.; Reilly, C.S.; Bitterman, P.; et al. Conserved GU-Rich Elements Mediate mRNA Decay by Binding to CUG-Binding Protein 1. Mol. Cell 2008, 29, 263–270. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Halees, A.; El-Badrawi, R.; Khabar, K.S.A. ARED Organism: Expansion of ARED reveals AU-rich element cluster variations between human and mouse. Nucleic Acids Res. 2007, 36, D137–D140. [Google Scholar] [CrossRef] [Green Version]
- Vasudevan, S.; Peltz, S.W. Regulated ARE-Mediated mRNA Decay in Saccharomyces cerevisiae. Mol. Cell 2001, 7, 1191–1200. [Google Scholar] [CrossRef]
- Lai, W.S.; Kennington, E.A.; Blackshear, P.J. Tristetraprolin and Its Family Members Can Promote the Cell-Free Deadenylation of AU-Rich Element-Containing mRNAs by Poly(A) Ribonuclease. Mol. Cell. Biol. 2003, 23, 3798–3812. [Google Scholar] [CrossRef] [Green Version]
- Gherzi, R.; Lee, K.-Y.; Briata, P.; Wegmüller, D.; Moroni, C.; Karin, M.; Chen, C.-Y. A KH Domain RNA Binding Protein, KSRP, Promotes ARE-Directed mRNA Turnover by Recruiting the Degradation Machinery. Mol. Cell 2004, 14, 571–583. [Google Scholar] [CrossRef]
- Barreau, C. AU-rich elements and associated factors: Are there unifying principles? Nucleic Acids Res. 2005, 33, 7138–7150. [Google Scholar] [CrossRef] [PubMed]
- Miyamoto, S.; Chiorini, J.A.; Urcelay, E.; Safer, B. Regulation of gene expression for translation initiation factor eIF-2α: Importance of the 3′ untranslated region. Biochem. J. 1996, 315, 791–798. [Google Scholar] [CrossRef] [PubMed]
- Costanzo, M.C.; Crawford, M.E.; Hirschman, J.E.; Kranz, J.E.; Olsen, P.; Robertson, L.S.; Skrzypek, M.S.; Braun, B.R.; Hopkins, K.L.; Kondu, P.; et al. YPDTM, PombePDTM and WormPDTM: Model organism volumes of the BioKnowledgeTM Library, an integrated resource for protein information. Nucleic Acids Res. 2001, 29, 75–79. [Google Scholar] [CrossRef] [Green Version]
- Scherrer, T.; Mittal, N.; Janga, S.C.; Gerber, A.P. A Screen for RNA-Binding Proteins in Yeast Indicates Dual Functions for Many Enzymes. PLoS ONE 2010, 5, e15499. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beckmann, B.; Horos, R.; Fischer, B.; Castello, A.; Eichelbaum, K.; Alleaume, A.-M.; Schwarzl, T.; Curk, T.; Foehr, S.; Huber, W.; et al. The RNA-binding proteomes from yeast to man harbour conserved enigmRBPs. Nat. Commun. 2015, 6, 10127. [Google Scholar] [CrossRef]
- Mitchell, S.F.; Jain, S.; She, M.; Parker, R. Global analysis of yeast mRNPs. Nat. Struct. Mol. Biol. 2012, 20, 127–133. [Google Scholar] [CrossRef] [Green Version]
- Matia-González, A.M.; Laing, E.E.; Gerber, A.P. Conserved mRNA-binding proteomes in eukaryotic organisms. Nat. Struct. Mol. Biol. 2015, 22, 1027–1033. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hogan, D.J.; Riordan, D.P.; Gerber, A.P.; Herschlag, D.; Brown, P.O. Diverse RNA-Binding Proteins Interact with Functionally Related Sets of RNAs, Suggesting an Extensive Regulatory System. PLoS Biol. 2008, 6, e255. [Google Scholar] [CrossRef] [Green Version]
- Wickens, M.; Bernstein, D.S.; Kimble, J.; Parker, R. A PUF family portrait: 3′UTR regulation as a way of life. Trends Genet. 2002, 18, 150–157. [Google Scholar] [CrossRef]
- Zamore, P.; Williamson, J.; Lehmann, R. The Pumilio protein binds RNA through a conserved domain that defines a new class of RNA-binding proteins. RNA 1997, 3, 1421–1433. [Google Scholar] [PubMed]
- Chagnovich, D.; Lehmann, R. Poly(A)-independent regulation of maternal hunchback translation in the Drosophila embryo. Proc. Natl. Acad. Sci. USA 2001, 98, 11359–11364. [Google Scholar] [CrossRef] [Green Version]
- Goldstrohm, A.C.; Hook, B.A.; Seay, D.J.; Wickens, M. PUF proteins bind Pop2p to regulate messenger RNAs. Nat. Struct. Mol. Biol. 2006, 13, 533–539. [Google Scholar] [CrossRef]
- Wreden, C.; Verrotti, A.; Schisa, J.; Lieberfarb, M.; Strickland, S. Nanos and pumilio establish embryonic polarity in Drosophila by promoting posterior deadenylation of hunchback mRNA. Development 1997, 124, 3015–3023. [Google Scholar] [CrossRef]
- Sonoda, J.; Wharton, R.P. Recruitment of Nanos to hunchback mRNA by Pumilio. Genes Dev. 1999, 13, 2704–2712. [Google Scholar] [CrossRef] [Green Version]
- Sonoda, J. Drosophila Brain Tumor is a translational repressor. Genes Dev. 2001, 15, 762–773. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barker, D.D.; Wang, C.; Moore, J.; Dickinson, L.K.; Lehmann, R. Pumilio is essential for function but not for distribution of the Drosophila abdominal determinant Nanos. Genes Dev. 1992, 6, 2312–2326. [Google Scholar] [CrossRef] [PubMed]
- Murata, Y.; Wharton, R.P. Binding of pumilio to maternal hunchback mRNA is required for posterior patterning in drosophila embryos. Cell 1995, 80, 747–756. [Google Scholar] [CrossRef] [Green Version]
- Gerber, A.P.; Herschlag, D.; Brown, P.O. Extensive Association of Functionally and Cytotopically Related mRNAs with Puf Family RNA-Binding Proteins in Yeast. PLoS Biol. 2004, 2, e79. [Google Scholar] [CrossRef] [PubMed]
- Olivas, W. The Puf3 protein is a transcript-specific regulator of mRNA degradation in yeast. EMBO J. 2000, 19, 6602–6611. [Google Scholar] [CrossRef] [Green Version]
- Fan, X.C.; Steitz, J.A. HNS, a nuclear-cytoplasmic shuttling sequence in HuR. Proc. Natl. Acad. Sci. USA 1998, 95, 15293–15298. [Google Scholar] [CrossRef] [Green Version]
- Peng, S.S.; Chen, C.A.; Xu, N.; Shyu, A. RNA stabilization by the AU-rich element binding protein, HuR, an ELAV protein. EMBO J. 1998, 17, 3461–3470. [Google Scholar] [CrossRef]
- Fan, X.C. Overexpression of HuR, a nuclear-cytoplasmic shuttling protein, increases the invivo stability of ARE-containing mRNAs. EMBO J. 1998, 17, 3448–3460. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Levy, N.S.; Chung, S.; Furneaux, H.; Levy, A.P. Hypoxic Stabilization of Vascular Endothelial Growth Factor mRNA by the RNA-binding Protein HuR. J. Biol. Chem. 1998, 273, 6417–6423. [Google Scholar] [CrossRef] [Green Version]
- Chang, N.; Yi, J.; Guo, G.; Liu, X.; Shang, Y.; Tong, T.; Cui, Q.; Zhan, M.; Gorospe, M.; Wang, W. HuR Uses AUF1 as a Cofactor to Promote p16 INK4 mRNA Decay. Mol. Cell. Biol. 2010, 30, 3875–3886. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, H.H.; Kuwano, Y.; Srikantan, S.; Lee, E.K.; Martindale, J.L.; Gorospe, M. HuR recruits let-7/RISC to repress c-Myc expression. Genes Dev. 2009, 23, 1743–1748. [Google Scholar] [CrossRef] [Green Version]
- Ramos-Alonso, L.; Romero, A.M.; Soler, M.; Perea-García, A.; Alepuz, P.; Puig, S.; Martínez-Pastor, M.T. Yeast Cth2 protein represses the translation of ARE-containing mRNAs in response to iron deficiency. PLoS Genet. 2018, 14, e1007476. [Google Scholar] [CrossRef] [Green Version]
- Timmusk, T.; Palm, K.; Metsis, M.; Reintam, T.; Paalme, V.; Saarma, M.; Persson, H. Multiple promoters direct tissue-specific expression of the rat BDNF gene. Neuron 1993, 10, 475–489. [Google Scholar] [CrossRef]
- An, J.J.; Gharami, K.; Liao, G.-Y.; Woo, N.H.; Lau, A.G.; Vanevski, F.; Torre, E.R.; Jones, K.; Feng, Y.; Lu, B.; et al. Distinct Role of Long 3′ UTR BDNF mRNA in Spine Morphology and Synaptic Plasticity in Hippocampal Neurons. Cell 2008, 134, 175–187. [Google Scholar] [CrossRef] [Green Version]
- Gonsalvez, G.B.; Urbinati, C.R.; Long, R.M. RNA localization in yeast: Moving towards a mechanism. Biol. Cell 2005, 97, 75–86. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shepard, K.A.; Gerber, A.P.; Jambhekar, A.; Takizawa, P.A.; Brown, P.O.; Herschlag, D.; DeRisi, J.L.; Vale, R.D. Widespread cytoplasmic mRNA transport in yeast: Identification of 22 bud-localized transcripts using DNA microarray analysis. Proc. Natl. Acad. Sci. USA 2003, 100, 11429–11434. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takizawa, P.A.; DeRisi, J.L.; Wilhelm, J.E.; Vale, R.D. Plasma Membrane Compartmentalization in Yeast by Messenger RNA Transport and a Septin Diffusion Barrier. Science 2000, 290, 341–344. [Google Scholar] [CrossRef] [Green Version]
- Takizawa, P.A.; Sil, A.; Swedlow, J.; Herskowitz, I.; Vale, R.D. Actin-dependent localization of an RNA encoding a cell-fate determinant in yeast. Nature 1997, 389, 90–93. [Google Scholar] [CrossRef]
- Jüschke, C.; Ferring, D.; Jansen, R.-P.; Seedorf, M. A Novel Transport Pathway for a Yeast Plasma Membrane Protein Encoded by a Localized mRNA. Curr. Biol. 2004, 14, 406–411. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Long, R.M.; Singer, R.H.; Meng, X.; Gonzalez, I.; Nasmyth, K.; Jansen, R.-P. Mating Type Switching in Yeast Controlled by Asymmetric Localization of ASH1 mRNA. Science 1997, 277, 383–387. [Google Scholar] [CrossRef] [Green Version]
- Long, R.M.; Gu, W.; Lorimer, E.; Singer, R.H.; Chartrand, P. She2p is a novel RNA-binding protein that recruits the Myo4p–She3p complex to ASH1 mRNA. EMBO J. 2000, 19, 6592–6601. [Google Scholar] [CrossRef] [Green Version]
- Munchow, S.; Sauter, C.; Jansen, R. Association of the class V myosin Myo4p with a localised messenger RNA in budding yeast depends on She proteins. J. Cell Sci. 1999, 112, 1511–1518. [Google Scholar] [CrossRef]
- Böhl, F.; Kruse, C.; Frank, A.; Ferring, D.; Jansen, R. She2p, a novel RNA-binding protein tethers ASH1 mRNA to the Myo4p myosin motor via She3p. EMBO J. 2000, 19, 5514–5524. [Google Scholar] [CrossRef] [Green Version]
- Takizawa, P.A.; Vale, R.D. The myosin motor, Myo4p, binds Ash1 mRNA via the adapter protein, She3p. Proc. Natl. Acad. Sci. USA 2000, 97, 5273–5278. [Google Scholar] [CrossRef] [Green Version]
- Chartrand, P.; Meng, X.-H.; Singer, R.; Long, R. Structural elements required for the localization of ASH1 mRNA and of a green fluorescent protein reporter particle in vivo. Curr. Biol. 1999, 9, 333–338. [Google Scholar] [CrossRef] [Green Version]
- Gonzalez, I.; Buonomo, S.; Nasmyth, K.; von Ahsen, U. ASH1 mRNA localization in yeast involves multiple secondary structural elementsand Ash1 protein translation. Curr. Biol. 1999, 9, 337–340. [Google Scholar] [CrossRef] [Green Version]
- Chartrand, P.; Meng, X.H.; Huttelmaier, S.; Donato, D.; Singer, R.H. Asymmetric Sorting of Ash1p in Yeast Results from Inhibition of Translation by Localization Elements in the mRNA. Mol. Cell 2002, 10, 1319–1330. [Google Scholar] [CrossRef]
- Gu, W.; Deng, Y.; Zenklusen, D.; Singer, R.H. A new yeast PUF family protein, Puf6p, represses ASH1 mRNA translation and is required for its localization. Genes Dev. 2004, 18, 1452–1465. [Google Scholar] [CrossRef] [Green Version]
- Corral-Debrinski, M.; Blugeon, C.; Jacq, C. In Yeast, the 3′ Untranslated Region or the Presequence of ATM1 Is Required for the Exclusive Localization of Its mRNA to the Vicinity of Mitochondria. Mol. Cell. Biol. 2000, 20, 7881–7892. [Google Scholar] [CrossRef]
- Marc, P.; Margeot, A.; Devaux, F.; Blugeon, C.; Corral-Debrinski, M.; Jacq, C. Genome-wide analysis of mRNAs targeted to yeast mitochondria. EMBO Rep. 2002, 3, 159–164. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Margeot, A.; Blugeon, C.; Sylvestre, J.; Vialette, S.; Jacq, C.; Corral-Debrinski, M. In Saccharomyces cerevisiae, ATP2 mRNA sorting to the vicinity of mitochondria is essential for respiratory function. EMBO J. 2002, 21, 6893–6904. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sylvestre, J.; Margeot, A.; Jacq, C.; Dujardin, G.; Corral-Debrinski, M. The Role of the 3′ Untranslated Region in mRNA Sorting to the Vicinity of Mitochondria Is Conserved from Yeast to Human Cells. Mol. Biol. Cell 2003, 14, 3848–3856. [Google Scholar] [CrossRef] [Green Version]
- Sylvestre, J.; Vialette, S.; Debrinski, M.C.; Jacq, C. Long mRNAs coding for yeast mitochondrial proteins of prokaryotic origin preferentially localize to the vicinity of mitochondria. Genome Biol. 2003, 4, R44. [Google Scholar] [CrossRef] [Green Version]
- Berkovits, B.D.; Mayr, C. Alternative 3′ UTRs act as scaffolds to regulate membrane protein localization. Nature 2015, 522, 363–367. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pinto, P.A.B.; Henriques, T.; Freitas, M.; Martins, T.; Domingues, R.G.; Wyrzykowska, P.S.; Coelho, P.A.; Carmo, A.; Sunkel, C.; Proudfoot, N.; et al. RNA polymerase II kinetics inpolopolyadenylation signal selection. EMBO J. 2011, 30, 2431–2444. [Google Scholar] [CrossRef] [Green Version]
- Dai, W.; Zhang, G.; Makeyev, E.V. RNA-binding protein HuR autoregulates its expression by promoting alternative polyadenylation site usage. Nucleic Acids Res. 2011, 40, 787–800. [Google Scholar] [CrossRef] [Green Version]
- Boutet, S.C.; Cheung, T.H.; Quach, N.L.; Liu, L.; Prescott, S.L.; Edalati, A.; Iori, K.; Rando, T.A. Alternative Polyadenylation Mediates MicroRNA Regulation of Muscle Stem Cell Function. Cell Stem Cell 2012, 10, 327–336. [Google Scholar] [CrossRef] [Green Version]
- Beilharz, T.; Humphreys, D.; Clancy, J.L.; Thermann, R.; Martin, D.I.K.; Hentze, M.; Preiss, T. microRNA-Mediated Messenger RNA Deadenylation Contributes to Translational Repression in Mammalian Cells. PLoS ONE 2009, 4, e6783. [Google Scholar] [CrossRef]
- Tranter, M.; Helsley, R.; Paulding, W.R.; McGuinness, M.; Brokamp, C.; Haar, L.; Liu, Y.; Ren, X.; Jones, W.K. Coordinated Post-transcriptional Regulation of Hsp70.3 Gene Expression by MicroRNA and Alternative Polyadenylation. J. Biol. Chem. 2011, 286, 29828–29837. [Google Scholar] [CrossRef] [Green Version]
- Akman, B.H.; Can, T.; Erson-Bensan, A.E. Estrogen-induced upregulation and 3′-UTR shortening of CDC6. Nucleic Acids Res. 2012, 40, 10679–10688. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hinske, L.C.; Galante, P.A.F.; Limbeck, E.; Möhnle, P.; Parmigiani, R.B.; Ohno-Machado, L.; Camargo, A.A.; Kreth, S. Alternative Polyadenylation Allows Differential Negative Feedback of Human miRNA miR-579 on Its Host Gene ZFR. PLoS ONE 2015, 10, e0121507. [Google Scholar] [CrossRef] [Green Version]
- Floor, S.N.; Doudna, J.A. Tunable protein synthesis by transcript isoforms in human cells. eLife 2016, 5, e10921. [Google Scholar] [CrossRef] [PubMed]
- Smith, L.; Coleman, L.J.; Cummings, M.; Satheesha, S.; Shaw, S.O.; Speirs, V.; Hughes, T.A. Expression of oestrogen receptor β isoforms is regulated by transcriptional and post-transcriptional mechanisms. Biochem. J. 2010, 429, 283–290. [Google Scholar] [CrossRef] [Green Version]
- Britten, R.J.; Roberts, R.B. High-Resolution Density Gradient Sedimentation Analysis. Science 1960, 131, 32–33. [Google Scholar] [CrossRef]
- Warner, J.R.; Knopf, P.M.; Rich, A. A multiple ribosomal structure in protein synthesis. Proc. Natl. Acad. Sci. USA 1963, 49, 122–129. [Google Scholar] [CrossRef] [Green Version]
- Arava, Y.; Wang, Y.; Storey, J.; Liu, C.L.; Brown, P.O.; Herschlag, D. Genome-wide analysis of mRNA translation profiles in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 2003, 100, 3889–3894. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, J.; Melton, C.; Suh, N.; Oh, J.S.; Horner, K.; Xie, F.; Sette, C.; Blelloch, R.; Conti, M. Genome-wide analysis of translation reveals a critical role for deleted in azoospermia-like (Dazl) at the oocyte-to-zygote transition. Genes Dev. 2011, 25, 755–766. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carrascoso, I.; Sánchez-Jiménez, C.; Izquierdo, J.M. Genome-wide profiling reveals a role for T-cell intracellular antigens TIA1 and TIAR in the control of translational specificity in HeLa cells. Biochem. J. 2014, 461, 43–50. [Google Scholar] [CrossRef] [PubMed]
- Larsson, O.; Tian, B.; Sonenberg, N. Toward a Genome-Wide Landscape of Translational Control. Cold Spring Harb. Perspect. Biol. 2012, 5, a012302. [Google Scholar] [CrossRef] [Green Version]
- Del Prete, M.J.; Vernal, R.; Dolznig, H.; Mullner, E.W.; Garcia-Sanz, J.A. Isolation of polysome-bound mRNA from solid tissues amenable for RT-PCR and profiling experiments. RNA 2007, 13, 414–421. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weidner, J.; Wang, C.; Prescianotto-Baschong, C.; Estrada, A.F.; Spang, A. The polysome-associated proteins Scp160 and Bfr1 prevent P body formation under normal growth conditions. J. Cell Sci. 2014, 127, 1992–2004. [Google Scholar] [CrossRef] [Green Version]
- Thermann, R.; Hentze, M.W. Drosophila miR2 induces pseudo-polysomes and inhibits translation initiation. Nature 2007, 447, 875–878. [Google Scholar] [CrossRef]
- Huch, S.; Gommlich, J.; Muppavarapu, M.; Beckham, C.; Nissan, T. Membrane-association of mRNA decapping factors is independent of stress in budding yeast. Sci. Rep. 2016, 6, 25477. [Google Scholar] [CrossRef] [Green Version]
- Ingolia, N.T.; Ghaemmaghami, S.; Newman, J.R.S.; Weissman, J.S. Genome-Wide Analysis in Vivo of Translation with Nucleotide Resolution Using Ribosome Profiling. Science 2009, 324, 218–223. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Archer, S.; Shirokikh, N.; Beilharz, T.; Preiss, T. Dynamics of ribosome scanning and recycling revealed by translation complex profiling. Nature 2016, 535, 570–574. [Google Scholar] [CrossRef] [PubMed]
- Steitz, J.A. Polypeptide Chain Initiation: Nucleotide Sequences of the Three Ribosomal Binding Sites in Bacteriophage R17 RNA. Nature 1969, 224, 957–964. [Google Scholar] [CrossRef]
- Yoshikawa, H.; Larance, M.; Harney, D.J.; Sundaramoorthy, R.; Ly, T.; Owen-Hughes, T.; Lamond, A.I. Efficient analysis of mammalian polysomes in cells and tissues using Ribo Mega-SEC. eLife 2018, 7, e36530. [Google Scholar] [CrossRef] [PubMed]
- Inada, T.; Winstall, E.; Tarun, S.Z.; Yates, J.R.; Schieltz, D.; Sachs, A.B. One-step affinity purification of the yeast ribosome and its associated proteins and mRNAs. RNA 2002, 8, 948–958. [Google Scholar] [CrossRef] [Green Version]
- Halbeisen, R.E.; Scherrer, T.; Gerber, A.P. Affinity purification of ribosomes to access the translatome. Methods 2009, 48, 306–310. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Turner, R.E.; Beilharz, T.H. Seeking a Role for Translational Control by Alternative Polyadenylation in Saccharomyces cerevisiae. Microorganisms 2021, 9, 1885. https://doi.org/10.3390/microorganisms9091885
Turner RE, Beilharz TH. Seeking a Role for Translational Control by Alternative Polyadenylation in Saccharomyces cerevisiae. Microorganisms. 2021; 9(9):1885. https://doi.org/10.3390/microorganisms9091885
Chicago/Turabian StyleTurner, Rachael E., and Traude H. Beilharz. 2021. "Seeking a Role for Translational Control by Alternative Polyadenylation in Saccharomyces cerevisiae" Microorganisms 9, no. 9: 1885. https://doi.org/10.3390/microorganisms9091885
APA StyleTurner, R. E., & Beilharz, T. H. (2021). Seeking a Role for Translational Control by Alternative Polyadenylation in Saccharomyces cerevisiae. Microorganisms, 9(9), 1885. https://doi.org/10.3390/microorganisms9091885




