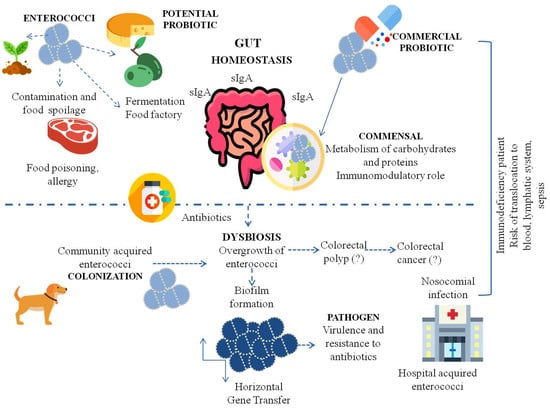
The Many Faces of Enterococcus spp.—Commensal, Probiotic and Opportunistic Pathogen
Abstract
:1. Introduction
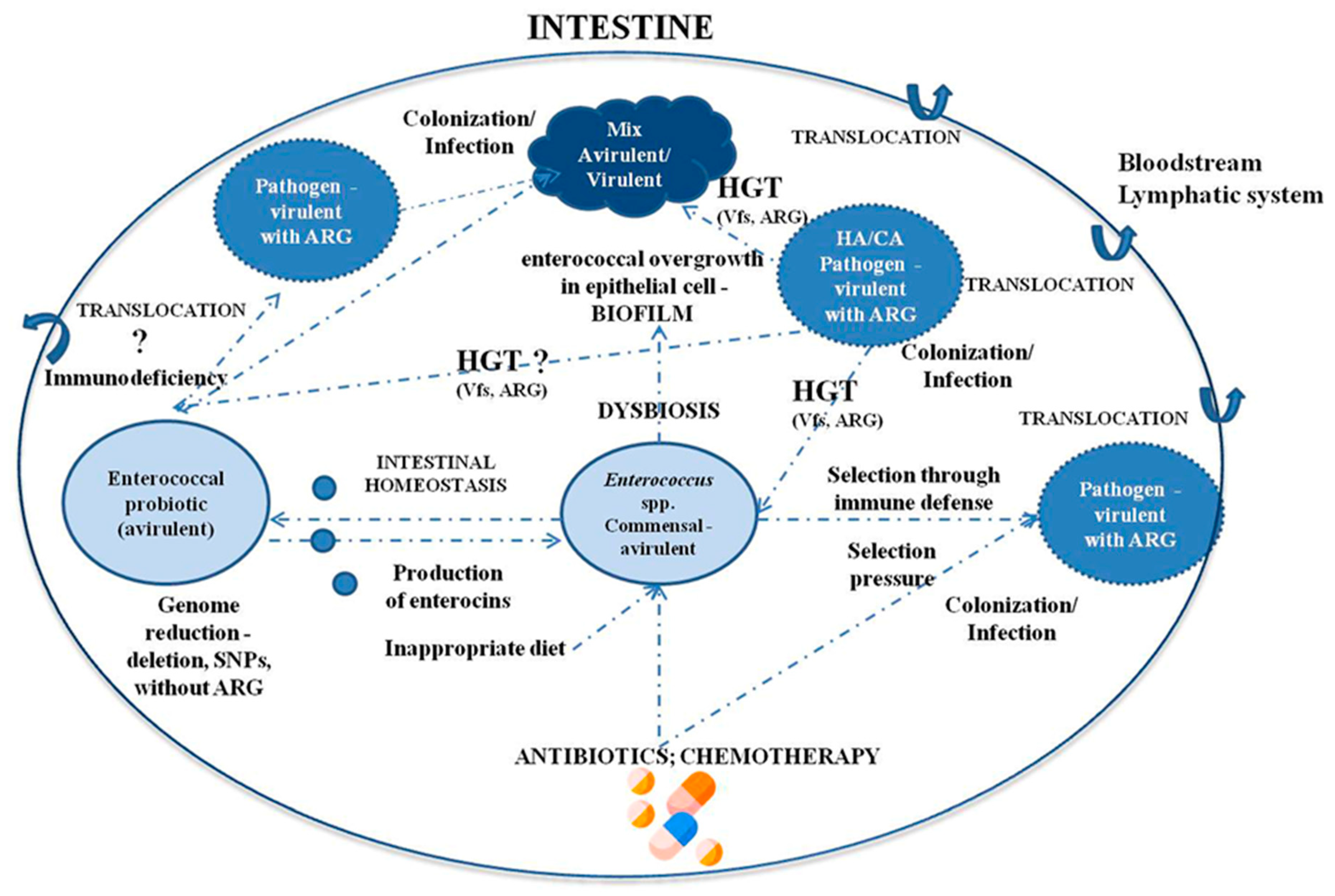
2. Enterococci as Commensal Microorganisms and Their Influence on the Immune System
3. Enterococci as Probiotics
3.1. Enterococcal Probiotic Strains
3.2. The Probiotic Importance of Enterococcus spp. and Applications
4. Enterococcus spp. as Opportunistic Pathogens
4.1. Hospital-Acquired Infection
4.2. Bacterial Translocation from the GI Tract to Organs
4.3. Mutagenic Effects and Theories of Tumorigenesis
4.4. Food-Borne Enterococci
5. Virulence Factors of Enterococcus spp. and Pathogenicity
6. The Problem of Antibiotic Resistance
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sadowy, E.; Luczkiewicz, A. Drug-resistant and hospital-associated Enterococcus faecium from wastewater, riverine estuary and anthropogenically impacted marine catchment basin. BMC Microbiol. 2014, 14, 66. [Google Scholar] [CrossRef] [Green Version]
- Martin, J.D.; Mundt, J.O. Enterococci in Insects. Appl. Microbiol. 1972, 24, 575–580. [Google Scholar] [CrossRef] [PubMed]
- Nowakiewicz, A.; Ziółkowska, G.; Trościańczyk, A.; Zięba, P.; Gnat, S. Determination of resistance and virulence genes in Enterococcus faecalis and E. faecium strains isolated from poultry and their genotypic characterization by ADSRRS-fingerprinting. Poult. Sci. 2017, 96, 986–996. [Google Scholar] [CrossRef]
- Micallef, S.A.; Rosenberg Goldstein, R.E.; George, A.; Ewing, L.; Tall, B.D.; Boyer, M.S.; Joseph, S.W.; Sapkota, A.R. Diversity, distribution and antibiotic resistance of Enterococcus spp. recovered from tomatoes, leaves, water and soil on U.S. Mid-Atlantic farms. Food Microbiol. 2013, 36, 465–474. [Google Scholar] [CrossRef]
- Abriouel, H.; Omar, N.B.; Molinos, A.C.; López, R.L.; Grande, M.J.; Martínez-Viedma, P.; Ortega, E.; Cañamero, M.M.; Galvez, A. Comparative analysis of genetic diversity and incidence of virulence factors and antibiotic resistance among enterococcal populations from raw fruit and vegetable foods, water and soil, and clinical samples. Int. J. Food Microbiol. 2008, 123, 38–49. [Google Scholar] [CrossRef]
- Franz, C.M.A.P.; Schillinger, U.; Holzapfel, W.H. Production and characterization of enterocin 900, a bacteriocin produced by Enterococcus faecium BFE 900 from black olives. Int. J. Food Microbiol. 1996, 29, 255–270. [Google Scholar] [CrossRef]
- Mundt, J.O. Occurrence of Enterococci on Plants in a Wild Environment. Appl. Microbiol. 1963, 11, 141–144. [Google Scholar] [CrossRef]
- Müller, T.; Ulrich, A.; Ott, E.M.; Müller, M. Identification of plant-associated enterococci. J. Appl. Microbiol. 2001, 91, 268–278. [Google Scholar] [CrossRef] [PubMed]
- Saillant, V.; Lipuma, D.; Ostyn, E.; Joubert, L.; Boussac, A.; Guerin, H.; Brandelet, G.; Arnoux, P.; Lechardeur, D. A novel enterococcus faecalis heme transport regulator (Fhtr) senses host heme to control its intracellular homeostasis. MBio 2021, 12, e03392-20. [Google Scholar] [CrossRef] [PubMed]
- Laissue, J.A.; Chappuis, B.B.; Müller, C.; Reubi, J.C.; Gebbers, J.O. The intestinal immune system and its relation to disease. Dig. Dis. 1993, 11, 298–312. [Google Scholar] [CrossRef] [PubMed]
- García-Díez, J.; Saraiva, C. Use of starter cultures in foods from animal origin to improve their safety. Int. J. Environ. Res. Public Health 2021, 18, 2544. [Google Scholar] [CrossRef]
- Gelsomino, R.; Vancanneyt, M.; Condon, S.; Swings, J.; Cogan, T.M. Enterococcal diversity in the environment of an Irish Cheddar-type cheesemaking factory. Int. J. Food Microbiol. 2001, 71, 177–188. [Google Scholar] [CrossRef]
- Franz, C.M.A.P.; Huch, M.; Abriouel, H.; Holzapfel, W.; Gálvez, A. Enterococci as probiotics and their implications in food safety. Int. J. Food Microbiol. 2011, 151, 125–140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giraffa, G. Functionality of enterococci in dairy products. Int. J. Food Microbiol. 2003, 88, 215–222. [Google Scholar] [CrossRef]
- Giraffa, G. Enterococci from foods. FEMS Microbiol. Rev. 2002, 26, 163–171. [Google Scholar] [CrossRef]
- O’Driscoll, T.; Crank, C.W. Vancomycin-resistant enterococcal infections: Epidemiology, clinical manifestations, and optimal management. Infect. Drug Resist. 2015, 8, 217–230. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guzman Prieto, A.M.; van Schaik, W.; Rogers, M.R.C.; Coque, T.M.; Baquero, F.; Corander, J.; Willems, R.J.L. Global emergence and dissemination of enterococci as nosocomial pathogens: Attack of the clones? Front. Microbiol. 2016, 7, 788. [Google Scholar] [CrossRef] [Green Version]
- Cheng, Y.; Ling, Z.; Li, L. The Intestinal Microbiota and Colorectal Cancer. Front. Immunol. 2020, 11, 3100. [Google Scholar] [CrossRef]
- Thomas, A.M.; Segata, N. Multiple levels of the unknown in microbiome research. BMC Biol. 2019, 17, 48. [Google Scholar] [CrossRef] [Green Version]
- Yang, X.; Xie, L.; Li, Y.; Wei, C. More than 9,000,000 unique genes in human gut bacterial community: Estimating gene numbers inside a human body. PLoS ONE 2009, 4, e6074. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Jia, H.; Cai, X.; Zhong, H.; Feng, Q.; Sunagawa, S.; Arumugam, M.; Kultima, J.R.; Prifti, E.; Nielsen, T.; et al. An integrated catalog of reference genes in the human gut microbiome. Nat. Biotechnol. 2014, 32, 834–841. [Google Scholar] [CrossRef] [PubMed]
- Qin, J.; Li, R.; Raes, J.; Arumugam, M.; Burgdorf, K.S.; Manichanh, C.; Nielsen, T.; Pons, N.; Levenez, F.; Yamada, T.; et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 2010, 464, 59–65. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Filippo, C.; Cavalieri, D.; Di Paola, M.; Ramazzotti, M.; Poullet, J.B.; Massart, S.; Collini, S.; Pieraccini, G.; Lionetti, P. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc. Natl. Acad. Sci. USA 2010, 107, 14691–14696. [Google Scholar] [CrossRef] [Green Version]
- Koenig, J.E.; Spor, A.; Scalfone, N.; Fricker, A.D.; Stombaugh, J.; Knight, R.; Angenent, L.T.; Ley, R.E. Succession of microbial consortia in the developing infant gut microbiome. Proc. Natl. Acad. Sci. USA 2011, 108, 4578–4585. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arumugam, M.; Raes, J.; Pelletier, E.; Le Paslier, D.; Yamada, T.; Mende, D.R.; Fernandes, G.R.; Tap, J.; Bruls, T.; Batto, J.M.; et al. Enterotypes of the human gut microbiome. Nature 2011, 473, 174–180. [Google Scholar] [CrossRef] [PubMed]
- Kurokawa, K.; Itoh, T.; Kuwahara, T.; Oshima, K.; Toh, H.; Toyoda, A.; Takami, H.; Morita, H.; Sharma, V.K.; Srivastava, T.P.; et al. Comparative metagenomics revealed commonly enriched gene sets in human gut microbiomes. DNA Res. 2007, 14, 169–181. [Google Scholar] [CrossRef] [PubMed]
- Tap, J.; Mondot, S.; Levenez, F.; Pelletier, E.; Caron, C.; Furet, J.P.; Ugarte, E.; Muñoz-Tamayo, R.; Paslier, D.L.E.; Nalin, R.; et al. Towards the human intestinal microbiota phylogenetic core. Environ. Microbiol. 2009, 11, 2574–2584. [Google Scholar] [CrossRef] [PubMed]
- Murray, B.E. The life and times of the enterococcus. Clin. Microbiol. Rev. 1990, 3, 46–65. [Google Scholar] [CrossRef]
- Chenoweth, C.; Schaberg, D. The epidemiology of enterococci. Eur. J. Clin. Microbiol. Infect. Dis. 1990, 9, 80–89. [Google Scholar] [CrossRef]
- Gilmore, M.; Clewell, D.; Courvalin, P.; Dunny, G. The Enterococci: Pathogenesis, Molecular Biology, and Antibiotic Resistance; Murray, B.E., Rice, L.B., Eds.; ASM Press: Washington, DC, USA, 2002; Volume 10, p. 439. [Google Scholar]
- Wan, L.Y.M.; Chen, Z.J.; Shah, N.P.; El-Nezami, H. Modulation of Intestinal Epithelial Defense Responses by Probiotic Bacteria. Crit. Rev. Food Sci. Nutr. 2016, 56, 2628–2641. [Google Scholar] [CrossRef] [PubMed]
- Mu, C.; Yang, Y.; Zhu, W. Gut microbiota: The brain peacekeeper. Front. Microbiol. 2016, 7, 345. [Google Scholar] [CrossRef] [Green Version]
- Gewolb, I.H.; Schwalbe, R.S.; Taciak, V.L.; Harrison, T.S.; Panigrahi, P. Stool microflora in extremely low birthweight infants. Arch. Dis. Child. Fetal Neonatal Ed. 1999, 80, F167. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Růžičková, M.; Vítězová, M.; Kushkevych, I. The characterization of Enterococcus genus: Resistance mechanisms and inflammatory bowel disease. Open Med. 2020, 15, 211–224. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fisher, K.; Phillips, C. The ecology, epidemiology and virulence of Enterococcus. Microbiology 2009, 155, 1749–1757. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McHugh, C.P.; Zhang, P.; Michalek, S.; Eleazer, P.D. pH required to kill Enterococcus faecalis in vitro. J. Endod. 2004, 30, 218–219. [Google Scholar] [CrossRef]
- Kramer, A.; Schwebke, I.; Kampf, G. How long do nosocomial pathogens persist on inanimate surfaces? A systematic review. BMC Infect. Dis. 2006, 6, 130. [Google Scholar] [CrossRef] [Green Version]
- Valdes, A.M.; Walter, J.; Segal, E.; Spector, T.D. Role of the gut microbiota in nutrition and health. BMJ 2018, 361, 36–44. [Google Scholar] [CrossRef] [Green Version]
- Silva, N.; Igrejas, G.; Gonçalves, A.; Poeta, P. Commensal gut bacteria: Distribution of Enterococcus species and prevalence of Escherichia coli phylogenetic groups in animals and humans in Portugal. Ann. Microbiol. 2012, 62, 449–459. [Google Scholar] [CrossRef]
- Krawczyk, B.; Lewandowski, K.; Bronk, M.; Samet, A.; Myjak, P.S.; Kur, J. Evaluation of a novel method based on amplification of DNA fragments surrounding rare restriction sites (ADSRRS fingerprinting) for typing strains of vancomycin-resistant Enterococcus faecium. J. Microbiol. Methods 2003, 52, 341–351. [Google Scholar] [CrossRef]
- Szemiako, K.; Krawczyk, B.; Samet, A.; Śledzińska, A.; Nowicki, B.; Nowicki, S.; Kur, J. A subset of two adherence systems, acute pro-inflammatory pap genes and invasion coding dra, fim, or sfa, increases the risk of Escherichia coli translocation to the bloodstream. Eur. J. Clin. Microbiol. Infect. Dis. 2013, 32, 1579–1582. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Samet, A.; Śledzińska, A.; Krawczyk, B.; Hellmann, A.; Nowicki, S.; Kur, J.; Nowicki, B. Leukemia and risk of recurrent Escherichia coli bacteremia: Genotyping implicates E. Coli translocation from the colon to the bloodstream. Eur. J. Clin. Microbiol. Infect. Dis. 2013, 32, 1393–1400. [Google Scholar] [CrossRef] [Green Version]
- Gibson, G.R.; Hutkins, R.; Sanders, M.E.; Prescott, S.L.; Reimer, R.A.; Salminen, S.J.; Scott, K.; Stanton, C.; Swanson, K.S.; Cani, P.D.; et al. Expert consensus document: The International Scientific Association for Probiotics and Prebiotics (ISAPP) consensus statement on the definition and scope of prebiotics. Nat. Rev. Gastroenterol. Hepatol. 2017, 14, 491–502. [Google Scholar] [CrossRef] [Green Version]
- Archambaud, C.; Derré-Bobillot, A.; Lapaque, N.; Rigottier-Gois, L.; Serror, P. Intestinal translocation of enterococci requires a threshold level of enterococcal overgrowth in the lumen. Sci. Rep. 2019, 9, 8926. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- CPG Sec. 689.100 Direct-Fed Microbial Products|FDA. Available online: https://www.fda.gov/regulatory-information/search-fda-guidance-documents/cpg-sec-689100-direct-fed-microbial-products (accessed on 5 July 2021).
- Bednorz, C.; Guenther, S.; Oelgeschläger, K.; Kinnemann, B.; Pieper, R.; Hartmann, S.; Tedin, K.; Semmler, T.; Neumann, K.; Schierack, P.; et al. Feeding the probiotic Enterococcus faecium strain NCIMB 10415 to piglets specifically reduces the number of Escherichia coli pathotypes that adhere to the gut mucosa. Appl. Environ. Microbiol. 2013, 79, 7896–7904. [Google Scholar] [CrossRef] [Green Version]
- Institute of Medicine. Ending the War Metaphor. The Changing Agenda for Unraveling the Host-Microbe Relationship: Workshop Summary; The National Academies Press: Washington, DC, USA, 2006. [Google Scholar] [CrossRef]
- Koutsoumanis, K.; Allende, A.; Alvarez-Ordóñez, A.; Bolton, D.; Bover-Cid, S.; Chemaly, M.; Davies, R.; De Cesare, A.; Hilbert, F.; Lindqvist, R.; et al. Update of the list of QPS-recommended biological agents intentionally added to food or feed as notified to EFSA 11: Suitability of taxonomic units notified to EFSA until September 2019. EFSA J. 2020, 18, 5965. [Google Scholar] [CrossRef] [Green Version]
- Rusch, K.; Rusch, V. Mikrobiologische Therapie Grundlagen und Praxis; Georg Thieme Verlag: New York, NY, USA, 2001. [Google Scholar]
- Habermann, W.; Zimmermann, K.; Skarabis, H.; Kunze, R.; Rusch, V. Reduction of acute relapses in patients with chronic recurrent hypertrophic sinusitis during treatment with a bacterial immunostimulant (Enterococcus faecalis bacteriae of human origin—A medical probiotic). Arzneimittel-Forschung/Drug Res. 2002, 52, 622–627. [Google Scholar] [CrossRef]
- Zhou, Y.; Liang, Y.; Lynch, K.H.; Dennis, J.J.; Wishart, D.S. PHAST: A Fast Phage Search Tool. Nucleic Acids Res. 2011, 39, W347–W352. [Google Scholar] [CrossRef]
- Fritzenwanker, M.; Kuenne, C.; Billion, A.; Hain, T.; Zimmermann, K.; Goesmann, A.; Chakraborty, T.; Domann, E. Complete genome sequence of the probiotic Enterococcus faecalis Symbioflor 1 clone DSM 16431. Genome Announc. 2013, 1, 165–177. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wassenaar, T.M.; Marzorati2, M.; Beimfohr, C.; Siegl, A.; Zimmermann, K. Survival of Probiotic E. coli and Ent. faecalis in the Human Host after Oral Intake: Results from in Vitro and in Vivo Studies. Adv. Biotechnol. Microbiol. 2017, 2, 1–5. [Google Scholar] [CrossRef]
- Domann, E.; Hain, T.; Ghai, R.; Billion, A.; Kuenne, C.; Zimmermann, K.; Chakraborty, T. Comparative genomic analysis for the presence of potential enterococcal virulence factors in the probiotic Enterococcus faecalis strain Symbioflor 1. Int. J. Med. Microbiol. 2007, 297, 533–539. [Google Scholar] [CrossRef]
- Baccouri, O.; Boukerb, A.M.; Farhat, L.B.; Zébré, A.; Zimmermann, K.; Domann, E.; Cambronel, M.; Barreau, M.; Maillot, O.; Rincé, I.; et al. Probiotic Potential and Safety Evaluation of Enterococcus faecalis OB14 and OB15, Isolated from Traditional Tunisian Testouri Cheese and Rigouta, Using Physiological and Genomic Analysis. Front. Microbiol. 2019, 10, 881. [Google Scholar] [CrossRef]
- Vebø, H.C.; Solheim, M.; Snipen, L.; Nes, I.F.; Brede, D.A. Comparative genomic analysis of pathogenic and probiotic Enterococcus faecalis isolates, and their transcriptional responses to growth in human urine. PLoS ONE 2010, 5, e12489. [Google Scholar] [CrossRef] [Green Version]
- Nami, Y.; Haghshenas, B.; Haghshenas, M.; Abdullah, N.; Khosroushahi, A.Y. The Prophylactic effect of probiotic Enterococcus lactis IW5 against different human cancer cells. Front. Microbiol. 2015, 6, 1317. [Google Scholar] [CrossRef] [Green Version]
- Adnan, M.; Patel, M.; Hadi, S. Functional and health promoting inherent attributes of Enterococcus hirae F2 as a novel probiotic isolated from the digestive tract of the freshwater fish Catla catla. PeerJ 2017, 5, e3085. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, B.; Evivie, S.E.; Jin, D.; Meng, Y.; Li, N.; Yan, F.; Huo, G.; Liu, F. Complete genome sequence of Enterococcus durans KLDS6.0933, a potential probiotic strain with high cholesterol removal ability. Gut Pathog. 2018, 10, 32. [Google Scholar] [CrossRef] [Green Version]
- Wopereis, H.; Oozeer, R.; Knipping, K.; Belzer, C.; Knol, J. The first thousand days—Intestinal microbiology of early life: Establishing a symbiosis. Pediatr. Allergy Immunol. 2014, 25, 428–438. [Google Scholar] [CrossRef] [Green Version]
- Ouwehand, A.C.; Salminen, S.; Isolauri, E. Probiotics: An overview of beneficial effects. Antonie Leeuwenhoek 2002, 82, 279–289. [Google Scholar] [CrossRef]
- Marteau, P.; Seksik, P.; Lepage, P.; Dore, J. Cellular and Physiological Effects of Probiotics and Prebiotics. Mini-Rev. Med. Chem. 2012, 4, 889–896. [Google Scholar] [CrossRef] [PubMed]
- Hlivak, P.; Odraska, J.; Ferencik, M.; Ebringer, L.; Jahnova, E.; Mikes, Z. One-year application of probiotic strain Enterococcus faecium M-74 decreases serum cholesterol levels. Bratisl. Lek. Listy 2005, 106, 67–72. [Google Scholar] [PubMed]
- Mikeš, Z.; Ferenčík, M.; Jahnová, E.; Ebringer, L.; Čižnár, I. Hypocholesterolemic and immunostimulatory effects of orally applied Enterococcus fœcium M-74 in man. Folia Microbiol. 1995, 40, 639–646. [Google Scholar] [CrossRef] [PubMed]
- Ebringer, L.; Ferenčík, M.; Lahitová, N.; Kačáni, L.; Michálková, D. Anti-mutagenic and immuno-stimulatory properties of lactic acid bacteria. World J. Microbiol. Biotechnol. 1995, 11, 294–298. [Google Scholar] [CrossRef]
- Mego, M.; Koncekova, R.; Mikuskova, E.; Drgona, L.; Ebringer, L.; Demitrovicova, L.; Nemova, I.; Trupl, J.; Mardiak, J.; Koza, I.; et al. Prevention of febrile neutropenia in cancer patients by probiotic strain Enterococcus faecium M-74. Phase II study. Support. Care Cancer 2006, 14, 285–290. [Google Scholar] [CrossRef] [PubMed]
- European Food Safety Authority (EFSA). Opinion of the Scientific Panel on additives and products or substances used in animal feed (FEEDAP) on the updating of the criteria used in the assessment of bacteria for resistance to antibiotics of human or veterinary importance. EFSA J. 2005, 3, 223. [Google Scholar] [CrossRef]
- Mitra, A.K.; Rabbani, G.H. A double-blind, controlled trial of bioflorin (Streptococcus faecium SF68) in adults with acute diarrhea due to Vibrio cholerae and enterotoxigenic Escherichia coli. Gastroenterology 1990, 99, 1149–1152. [Google Scholar] [CrossRef]
- Bybee, S.N.; Scorza, A.V.; Lappin, M.R. Effect of the probiotic Enterococcus faecium SF68 on presence of diarrhea in cats and dogs housed in an animal shelter. J. Vet. Intern. Med. 2011, 25, 856–860. [Google Scholar] [CrossRef] [PubMed]
- Torres-Henderson, C.; Summers, S.; Suchodolski, J.; Lappin, M.R. Effect of Enterococcus Faecium Strain SF68 on Gastrointestinal Signs and Fecal Microbiome in Cats Administered Amoxicillin-Clavulanate. Top. Companion Anim. Med. 2017, 32, 104–108. [Google Scholar] [CrossRef]
- Al-Balawi, M.; Morsy, F.M. Enterococcus faecalis Is a Better Competitor Than Other Lactic Acid Bacteria in the Initial Colonization of Colon of Healthy Newborn Babies at First Week of Their Life. Front. Microbiol. 2020, 11, 2017. [Google Scholar] [CrossRef] [PubMed]
- Giraffa, G. Enterococcus. In Encyclopedia of Food Microbiology II Enterococcus; Elsevier: Amsterdam, The Netherlands, 2014; pp. 674–679. [Google Scholar] [CrossRef]
- Barrangou, R.; Lahtinen, S.J.; Ibrahim, F.; Ouwehand, A.C. Chapter 5: Genus Lactobacillus. In Lactic Acid Bacteria: Microbiological and Functional Aspects; Lahtinen, S., Ouwehand, A.C., Salminen, S., von Wright, A., Eds.; CRC Press: Boca Raton, FL, USA, 2012; ISBN 9781439836774. [Google Scholar]
- Abanoz, H.S.; Kunduhoglu, B. Antimicrobial activity of a bacteriocin produced by enterococcus faecalis kt11 against some pathogens and antibiotic-resistant Bacteria. Korean J. Food Sci. Anim. Resour. 2018, 38, 1064–1079. [Google Scholar] [CrossRef] [Green Version]
- Wu, Y.; Zhen, W.; Geng, Y.; Wang, Z.; Guo, Y. Effects of dietary Enterococcus faecium NCIMB 11181 supplementation on growth performance and cellular and humoral immune responses in broiler chickens. Poult. Sci. 2019, 98, 150–163. [Google Scholar] [CrossRef] [PubMed]
- Viaud, S.; Saccheri, F.; Mignot, G.; Yamazaki, T.; Daillère, R.; Hannani, D.; Enot, D.P.; Pfirschke, C.; Engblom, C.; Pittet, M.J.; et al. The intestinal microbiota modulates the anticancer immune effects of cyclophosphamide. Science 2013, 342, 971–976. [Google Scholar] [CrossRef] [Green Version]
- Benyacoub, J.; Czarnecki-Maulden, G.L.; Cavadini, C.; Sauthier, T.; Anderson, R.E.; Schiffrin, E.J.; Von der Weid, T. von Supplementation of food with Enterococcus faecium (SF68) stimulates immune functions in young dogs. J. Nutr. 2003, 133, 1158–1162. [Google Scholar] [CrossRef] [Green Version]
- Vahjen, W.; Taras, D.; Simon, O. Effect of the probiotic Enterococcus faecium NCIMB10415 on cell numbers of total Enterococcus spp., E. faecium and E. faecalis in the intestine of piglets. Curr. Issues Intest. Microbiol. 2007, 8, 1–8. [Google Scholar]
- Lucena-Padrós, H.; González, J.M.; Caballero-Guerrero, B.; Ruiz-Barba, J.L.; Maldonado-Barragán, A. Enterococcus olivae sp. nov., isolated from Spanish-style green-olive fermentations. Int. J. Syst. Evol. Microbiol. 2014, 64, 2534–2539. [Google Scholar] [CrossRef]
- Bhardwaj, A.; Malik, R.K.; Chauhan, P. Functional and safety aspects of enterococci in dairy foods. Indian J. Microbiol. 2008, 48, 317–325. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lebreton, F.; Willems, R.J.L.; Gilmore, M.S. Enterococcus Diversity, Origins in Nature, and Gut Colonization. In Enterococci: From Commensals to Leading Causes of Drug Resistant Infection [Internet]; 2014. Available online: https://www.ncbi.nlm.nih.gov/books/NBK190427/ (accessed on 1 September 2021).
- John, U.V.; Carvalho, J. Enterococcus: Review of its physiology, pathogenesis, diseases and the challenges it poses for clinical microbiology. Front. Biol. 2011, 6, 357–366. [Google Scholar]
- Kao, P.H.N.; Kline, K.A. Dr. Jekyll and Mr. Hide: How Enterococcus faecalis Subverts the Host Immune Response to Cause Infection. J. Mol. Biol. 2019, 431, 2932–2945. [Google Scholar] [CrossRef]
- Krawczyk, B.; Wysocka, M.; Kotłowski, R.; Bronk, M.; Michalik, M.; Samet, A. Linezolid-resistant Enterococcus faecium strains isolated from one hospital in Poland-commensals or hospital-adapted pathogens? PLoS ONE 2020, 15, e0233504. [Google Scholar] [CrossRef] [PubMed]
- Tendolkar, P.M.; Baghdayan, A.S.; Shankar, N. Pathogenic enterococci: New developments in the 21st century. Cell. Mol. Life Sci. 2003, 60, 2622–2636. [Google Scholar] [CrossRef]
- Krawczyk, B.; Samet, A.; Bronk, M.; Hellmann, A.; Kur, J. Emerging linezolid-resistant, vancomycin resistant Enterococcus faecium from a patient of a haematological unit in Poland. Pol. J. Microbiol. 2004, 53, 193–196. [Google Scholar] [PubMed]
- Fanaro, S.; Chierici, R.; Guerrini, P.; Vigi, V. Intestinal microflora in early infancy: Composition and development. Acta Paediatr. Int. J. Paediatr. Suppl. 2003, 91, 48–55. [Google Scholar] [CrossRef] [PubMed]
- Bretón, J.R.; Peset, V.; Morcillo, F.; Cano, J.; Sarrión, A.; Pérez-Belles, C.; Gobernado, M. Neonatal meningitis due to Enterococcus spp.: Presentation of four cases. Enferm. Infecc. Microbiol. Clin. 2002, 20, 443–447. [Google Scholar] [CrossRef]
- Montealegre, M.C.; Roh, J.H.; Rae, M.; Davlieva, M.G.; Singh, K.V.; Shamoo, Y.; Murray, B.E. Differential Penicillin-Binding Protein 5 (PBP5) levels in the enterococcus faecium clades with different levels of ampicillin resistance. Antimicrob. Agents Chemother. 2017, 61, e02034-16. [Google Scholar] [CrossRef] [Green Version]
- Correa-Martinez, C.L.; Tönnies, H.; Froböse, N.J.; Mellmann, A.; Kampmeier, S. Transmission of vancomycin-resistant enterococci in the hospital setting: Uncovering the patient–environment interplay. Microorganisms 2020, 8, 203. [Google Scholar] [CrossRef] [Green Version]
- O’Brien, V.P.; Hannan, T.J.; Nielsen, H.V.; Hultgren, S.J. Drug and Vaccine Development for the Treatment and Prevention of Urinary Tract Infections. Microbiol. Spectr. 2016, 4. [Google Scholar] [CrossRef] [Green Version]
- Armbruster, C.E.; Prenovost, K.; Mobley, H.L.T.; Mody, L. How Often Do Clinically Diagnosed Catheter-Associated Urinary Tract Infections in Nursing Homes Meet Standardized Criteria? J. Am. Geriatr. Soc. 2017, 65, 395–401. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Learman, B.S.; Brauer, A.L.; Eaton, K.A.; Armbruster, C.E. A rare opportunist, Morganella morganii, decreases severity of polymicrobial catheter-associated urinary tract infection. Infect. Immun. 2020, 88, e00691-19. [Google Scholar] [CrossRef] [PubMed]
- Wells, C.L.; Jechorek, R.P.; Gillingham, K.J. Relative Contributions of Host and Microbial Factors in Bacterial Translocation. Arch. Surg. 1991, 126, 247–252. [Google Scholar] [CrossRef]
- Fine, R.L.; Vieira, S.M.; Gilmore, M.S.; Kriegel, M.A. Mechanisms and consequences of gut commensal translocation in chronic diseases. Gut Microbes 2020, 11, 217. [Google Scholar] [CrossRef] [PubMed]
- Knoop, K.A.; McDonald, K.G.; Kulkarni, D.H.; Newberry, R.D. Antibiotics promote inflammation through the translocation of native commensal colonic bacteria. Gut 2016, 65, 1100–1109. [Google Scholar] [CrossRef] [Green Version]
- Diehl, G.E.; Longman, R.S.; Zhang, J.X.; Breart, B.; Galan, C.; Cuesta, A.; Schwab, S.R.; Littman, D.R. Microbiota restricts trafficking of bacteria to mesenteric lymph nodes by CX 3 CR1 hi cells. Nature 2013, 494, 116–120. [Google Scholar] [CrossRef]
- Zeng, J.; Teng, F.; Weinstock, G.M.; Murray, B.E. Translocation of Enterococcus faecalis Strains across a Monolayer of Polarized Human Enterocyte-Like T84 Cells. J. Clin. Microbiol. 2004, 42, 1149–1154. [Google Scholar] [CrossRef] [Green Version]
- Zeng, J.; Teng, F.; Murray, B.E. Gelatinase is important for translocation of Enterococcus faecalis across polarized human enterocyte-like T84 cells. Infect. Immun. 2005, 73, 1606–1612. [Google Scholar] [CrossRef] [Green Version]
- Manfredo Vieira, S.; Hiltensperger, M.; Kumar, V.; Zegarra-Ruiz, D.; Dehner, C.; Khan, N.; Costa, F.R.C.; Tiniakou, E.; Greiling, T.; Ruff, W.; et al. Translocation of a gut pathobiont drives autoimmunity in mice and humans. Science 2018, 359, 1156–1161. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eccles, L.J.; O’Neill, P.; Lomax, M.E. Delayed repair of radiation induced clustered DNA damage: Friend or foe? Mutat. Res.—Fundam. Mol. Mech. Mutagen. 2011, 711, 134–141. [Google Scholar] [CrossRef]
- Escolà-Vergé, L.; Peghin, M.; Givone, F.; Pérez-Rodríguez, M.T.; Suárez-Varela, M.; Meije, Y.; Abelenda, G.; Almirante, B.; Fernández-Hidalgo, N. Prevalence of colorectal disease in Enterococcus faecalis infective endocarditis: Results of an observational multicenter study. Rev. Esp. Cardiol. (Engl. Ed.) 2020, 73, 711–717. [Google Scholar] [CrossRef] [PubMed]
- Frickmann, H.; Köller, K.; Veil, I.; Weise, M.; Ludyga, A.; Schwarz, N.G.; Warnke, P.; Podbielski, A. On the role of enterococci in the bloodstream: Results of a single-center, retrospective, observational study at a German University Hospital. Eur. J. Microbiol. Immunol. 2017, 7, 284–295. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Allen, T.D.; May, R.J.; Lightfoot, S.; Houchen, C.W.; Huycke, M.M. Enterococcus faecalis induces aneuploidy and tetraploidy in colonic epithelial cells through a bystander effect. Cancer Res. 2008, 68, 9909–9917. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rezasoltani, S.; Asadzadeh Aghdaei, H.; Dabiri, H.; Akhavan Sepahi, A.; Modarressi, M.H.; Nazemalhosseini Mojarad, E. The association between fecal microbiota and different types of colorectal polyp as precursors of colorectal cancer. Microb. Pathog. 2018, 124, 244–249. [Google Scholar] [CrossRef]
- de Andrade Calaça, P.R.; da Silva Santos, D.; da Silva, J.F.; Aragão, A.B.L.; de Melo, I.M.F.; da Silva, E.C.S.; Porto, A.L.F.; Soares, M.T.C.V. Enterococcus faecium 137v como fator de proteção em modelo animal para câncer colorretal. Res. Soc. Dev. 2021, 10, e9110615354. [Google Scholar] [CrossRef]
- Wang, T.; Cai, G.; Qiu, Y.; Fei, N.; Zhang, M.; Pang, X.; Jia, W.; Cai, S.; Zhao, L. Structural segregation of gut microbiota between colorectal cancer patients and healthy volunteers. ISME J. 2012, 6, 320–329. [Google Scholar] [CrossRef] [Green Version]
- Huycke, M.M.; Abrams, V.; Moore, D.R. Enterococcus faecalis produces extracellular superoxide and hydrogen peroxide that damages colonic epithelial cell DNA. Carcinogenesis 2002, 23, 529–536. [Google Scholar] [CrossRef] [PubMed]
- Allen, T.D.; Moore, D.R.; Wang, X.; Casu, V.; May, R.; Lerner, M.R.; Houchen, C.; Brackett, D.J.; Huycke, M.M. Dichotomous metabolism of Enterococcus faecalis induced by haematin starvation modulates colonic gene expression. J. Med. Microbiol. 2008, 57, 1193–1204. [Google Scholar] [CrossRef]
- Evans, M.D.; Dizdaroglu, M.; Cooke, M.S. Oxidative DNA damage and disease: Induction, repair and significance. Mutat. Res.—Rev. Mutat. Res. 2004, 567, 1–61. [Google Scholar] [CrossRef] [PubMed]
- Huycke, M.M.; Moore, D.R. In vivo production of hydroxyl radical by Enterococcus faecalis colonizing the intestinal tract using aromatic hydroxylation. Free Radic. Biol. Med. 2002, 33, 818–826. [Google Scholar] [CrossRef]
- Grootaert, C.; Van de Wiele, T.; Van Roosbroeck, I.; Possemiers, S.; Vercoutter-Edouart, A.S.; Verstraete, W.; Bracke, M.; Vanhoecke, B. Bacterial monocultures, propionate, butyrate and H2O2 modulate the expression, secretion and structure of the fasting-induced adipose factor in gut epithelial cell lines. Environ. Microbiol. 2011, 13, 1778–1789. [Google Scholar] [CrossRef]
- Fracalanzza, S.A.P.; Scheidegger, E.M.D.; Dos Santos, P.F.; Leite, P.C.; Teixeira, L.M. Antimicrobial resistance profiles of enterococci isolated from poultry meat and pasteurized milk in Rio de Janeiro, Brazil. Mem. Inst. Oswaldo Cruz 2007, 102, 853–859. [Google Scholar] [CrossRef] [Green Version]
- Bortolaia, V.; Espinosa-Gongora, C.; Guardabassi, L. Human health risks associated with antimicrobial-resistant enterococci and Staphylococcus aureus on poultry meat. Clin. Microbiol. Infect. 2016, 22, 130–140. [Google Scholar] [CrossRef] [Green Version]
- Miranda, J.M.; Guarddon, M.; Mondragón, A.; Vázquez, B.I.; Fente, C.A.; Cepeda, A.; Franco, C.M. Antimicrobial resistance in Enterococcus spp. strains isolated from organic chicken, conventional chicken, and turkey meat: A comparative survey. J. Food Prot. 2007, 70, 1021–1024. [Google Scholar] [CrossRef]
- Chajęcka-Wierzchowska, W.; Zadernowska, A.; Zarzecka, U.; Zakrzewski, A.; Gajewska, J. Enterococci from ready-to-eat food—Horizontal gene transfer of antibiotic resistance genes and genotypic characterization by PCR melting profile. J. Sci. Food Agric. 2019, 99, 1172–1179. [Google Scholar] [CrossRef]
- Choi, J.M.; Woo, G.J. Transfer of Tetracycline Resistance Genes with Aggregation Substance in Food-Borne Enterococcus faecalis. Curr. Microbiol. 2015, 70, 476–484. [Google Scholar] [CrossRef] [Green Version]
- Haug, M.C.; Tanner, S.A.; Lacroix, C.; Stevens, M.J.A.; Meile, L. Monitoring horizontal antibiotic resistance gene transfer in a colonic fermentation model. FEMS Microbiol. Ecol. 2011, 78, 210–219. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hugas, M.; Garriga, M.; Aymerich, M.T. Functionalty of enterococci in meat products. Int. J. Food Microbiol. 2004, 88, 223–233. [Google Scholar] [CrossRef]
- Gaca, A.O.; Lemos, J.A. Adaptation to Adversity: The Intermingling of Stress Tolerance and Pathogenesis in Enterococci. Microbiol. Mol. Biol. Rev. 2019, 83, e00008-19. [Google Scholar] [CrossRef] [PubMed]
- Gilmore, M.S.; Coburn, P.S.; Nallapareddy, S.R.; Murray, B.E. Enterococcal Virulence. Enterococci 2014, 301–354. [Google Scholar] [CrossRef]
- Rich, R.L.; Kreikemeyer, B.; Owens, R.T.; LaBrenz, S.; Narayana, S.V.L.; Weinstock, G.M.; Murray, B.E.; Höök, M. Ace is a collagen-binding MSCRAMM from Enterococcus faecalis. J. Biol. Chem. 1999, 274, 26939–26945. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nallapareddy, S.R.; Weinstock, G.M.; Murray, B.E. Clinical isolates of Enterococcus faecium exhibit strain-specific collagen binding mediated by Acm, a new member of the MSCRAMM family. Mol. Microbiol. 2003, 47, 1733–1747. [Google Scholar] [CrossRef]
- de Freitas Silva, E.C.; Montalvão, C.R.; Bonafé, S. Infectious Endocarditis from Enterococcus faecalis Associated with Tubular Adenoma of the Sigmoid Colon. Case Rep. Infect. Dis. 2017, 2017, 3095031. [Google Scholar] [CrossRef] [Green Version]
- Nallapareddy, S.R.; Singh, K.V.; Okhuysen, P.C.; Murray, B.E. A functional collagen adhesin gene, acm, in clinical isolates of Enterococcus faecium correlates with the recent success of this emerging nosocomial pathogen. Infect. Immun. 2008, 76, 4110–4119. [Google Scholar] [CrossRef] [Green Version]
- Torabinejad, M.; Eby, W.C.; Naidorf, I.J. Inflammatory and immunological aspects of the pathogenesis of human periapical lesions. J. Endod. 1985, 11, 479–488. [Google Scholar] [CrossRef]
- Galli, D.; Friesenegger, A.; Wirth, R. Transcriptional control of sex-pheromone-inducible genes on plasmid pAD1 of Enterococcus faecalis and sequence analysis of a third structural gene for (pPD1-encoded) aggregation substance. Mol. Microbiol. 1992, 6, 1297–1308. [Google Scholar] [CrossRef]
- Kao, S.M.; Olmsted, S.B.; Viksnins, A.S.; Gallo, J.C.; Dunny, G.M. Molecular and genetic analysis of a region of plasmid pCF10 containing positive control genes and structural genes encoding surface proteins involved in pheromone-inducible conjugation in Enterococcus faecalis. J. Bacteriol. 1991, 173, 7650–7664. [Google Scholar] [CrossRef] [Green Version]
- Hendrickx, A.P.A.; Willems, R.J.L.; Bonten, M.J.M.; van Schaik, W. LPxTG surface proteins of enterococci. Trends Microbiol. 2009, 17, 423–430. [Google Scholar] [CrossRef]
- Shankar, V.; Baghdayan, A.S.; Huycke, M.M.; Lindahl, G.; Gilmore, M.S. Infection-derived Enterococcus faecalis strains are enriched in esp, a gene encoding a novel surface protein. Infect. Immun. 1999, 67, 193–200. [Google Scholar] [CrossRef] [Green Version]
- Mohamed, J.A.; Huang, D.B. Biofilm formation by enterococci. J. Med. Microbiol. 2007, 56, 1581–1588. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van Schaik, W.; Top, J.; Riley, D.R.; Boekhorst, J.; Vrijenhoek, J.E.P.; Schapendonk, C.M.E.; Hendrickx, A.P.A.; Nijman, I.J.; Bonten, M.J.M.; Tettelin, H.; et al. Pyrosequencing-based comparative genome analysis of the nosocomial pathogen Enterococcus faecium and identification of a large transferable pathogenicity island. BMC Genom. 2010, 11, 239. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Low, Y.L.; Jakubovics, N.S.; Flatman, J.C.; Jenkinson, H.F.; Smith, A.W. Manganese-dependent regulation of the endocarditis-associated virulence factor EfaA of Enterococcus faecafis. J. Med. Microbiol. 2003, 52, 113–119. [Google Scholar] [CrossRef] [PubMed]
- Nallapareddy, S.R.; Singh, K.V.; Sillanpää, J.; Garsin, D.A.; Höök, M.; Erlandsen, S.L.; Murray, B.E. Endocarditis and biofilm-associated pili of Enterococcus faecalis. J. Clin. Investig. 2006, 116, 2799–2807. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mundy, L.M.; Sahm, D.F.; Gilmore, M. Relationships between Enterococcal Virulence and Antimicrobial Resistance. Clin. Microbiol. Rev. 2000, 13, 513–522. [Google Scholar] [CrossRef]
- Del Papa, M.F.; Hancock, L.E.; Thomas, V.C.; Perego, M. Full activation of Enterococcus faecalis gelatinase by a C-terminal proteolytic cleavage. J. Bacteriol. 2007, 189, 8835–8843. [Google Scholar] [CrossRef] [Green Version]
- Thurlow, L.R.; Thomas, V.C.; Narayanan, S.; Olson, S.; Fleming, S.D.; Hancock, L.E. Gelatinase contributes to the pathogenesis of endocarditis caused by Enterococcus faecalis. Infect. Immun. 2010, 78, 4936–4943. [Google Scholar] [CrossRef] [Green Version]
- Engelbert, M.; Mylonakis, E.; Ausubel, F.M.; Calderwood, S.B.; Gilmore, M.S. Contribution of gelatinase, serine protease, and fsr to the pathogenesis of Enterococcus faecalis endophthalmitis. Infect. Immun. 2004, 72, 3628–3633. [Google Scholar] [CrossRef] [Green Version]
- Ali, L.; Goraya, M.U.; Arafat, Y.; Ajmal, M.; Chen, J.L.; Yu, D. Molecular mechanism of quorum-sensing in Enterococcus faecalis: Its role in virulence and therapeutic approaches. Int. J. Mol. Sci. 2017, 18, 960. [Google Scholar] [CrossRef] [Green Version]
- Kayaoglu, G.; Ørstavik, D. Virulence factors of Enterococcus faecalis: Relationship to endodontic disease. Crit. Rev. Oral Biol. Med. 2004, 15, 308–320. [Google Scholar] [CrossRef] [Green Version]
- Huycke, M.M.; Spiegel, C.A.; Gilmore, M.S. Bacteremia caused by hemolytic, high-level gentamicin-resistant Enterococcus faecalis. Antimicrob. Agents Chemother. 1991, 35, 1626–1634. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dapkevicius, M.d.L.E.; Sgardioli, B.; Câmara, S.P.A.; Poeta, P.; Malcata, F.X. Current trends of enterococci in dairy products: A comprehensive review of their multiple roles. Foods 2021, 10, 821. [Google Scholar] [CrossRef]
- İspirli, H.; Demirbaş, F.; Dertli, E. Characterization of functional properties of Enterococcus spp. isolated from Turkish white cheese. LWT—Food Sci. Technol. 2017, 75, 358–365. [Google Scholar] [CrossRef]
- Nieto-Arribas, P.; Seseña, S.; Poveda, J.M.; Chicón, R.; Cabezas, L.; Palop, L. Enterococcus populations in artisanal Manchego cheese: Biodiversity, technological and safety aspects. Food Microbiol. 2011, 28, 891–899. [Google Scholar] [CrossRef] [PubMed]
- Fuka, M.M.; Maksimovic, A.Z.; Tanuwidjaja, I.; Hulak, N.; Schloter, M. Characterization of enterococcal community isolated from an Artisan Istrian raw milk cheese: Biotechnologicaland safety aspects. Food Technol. Biotechnol. 2017, 55, 368–380. [Google Scholar] [CrossRef]
- Özkan, E.R.; Demirci, T.; Akın, N. In vitro assessment of probiotic and virulence potential of Enterococcus faecium strains derived from artisanal goatskin casing Tulum cheeses produced in central Taurus Mountains of Turkey. LWT 2021, 141, 110908. [Google Scholar] [CrossRef]
- McBride, S.M.; Coburn, P.S.; Baghdayan, A.S.; Willems, R.J.L.; Grande, M.J.; Shankar, N.; Gilmore, M.S. Genetic variation and evolution of the pathogenicity island of Enterococcus faecalis. J. Bacteriol. 2009, 191, 3392–3402. [Google Scholar] [CrossRef] [Green Version]
- Hacker, J.; Kaper, J.B. Pathogenicity islands and the evolution of microbes. Annu. Rev. Microbiol. 2000, 54, 641–679. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sparo, M.; Urbizu, L.; Solana, M.V.; Pourcel, G.; Delpech, G.; Confalonieri, A.; Ceci, M.; Sánchez Bruni, S.F. High-level resistance to gentamicin: Genetic transfer between Enterococcus faecalis isolated from food of animal origin and human microbiota. Lett. Appl. Microbiol. 2012, 54, 119–125. [Google Scholar] [CrossRef]
- Vignaroli, C.; Zandri, G.; Aquilanti, L.; Pasquaroli, S.; Biavasco, F. Multidrug-resistant enterococci in animal meat and faeces and Co-transfer of resistance from an Enterococcus durans to a human Enterococcus faecium. Curr. Microbiol. 2011, 62, 1438–1447. [Google Scholar] [CrossRef] [PubMed]
- Agersø, Y.; Pedersen, A.G.; Aarestrup, F.M. Identification of Tn5397-like and Tn916-like transposons and diversity of the tetracycline resistance gene tet(M) in enterococci from humans, pigs and poultry. J. Antimicrob. Chemother. 2006, 57, 832–839. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramos, Y.; Morales, D.K. Exopolysaccharide-mediated surface penetration as new virulence trait in Enterococcus faecalis. Commun. Integr. Biol. 2019, 12, 144–147. [Google Scholar] [CrossRef] [Green Version]
- Ebrahimi, A.; Schwartzman, J.; Cordero, O.X. Multicellular behaviour enables cooperation in microbial cell aggregates. Philos. Trans. R. Soc. B Biol. Sci. 2019, 374, 20190077. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lynch, K.M.; Zannini, E.; Coffey, A.; Arendt, E.K. Lactic Acid Bacteria Exopolysaccharides in Foods and Beverages: Isolation, Properties, Characterization, and Health Benefits. Annu. Rev. Food Sci. Technol. 2018, 9, 155–176. [Google Scholar] [CrossRef]
- Salachna, P.; Mizielińska, M.; Soból, M. Exopolysaccharide gellan gum and derived oligo-gellan enhance growth and antimicrobial activity in eucomis plants. Polymers 2018, 10, 242. [Google Scholar] [CrossRef] [Green Version]
- Abdalla, A.K.; Ayyash, M.M.; Olaimat, A.N.; Osaili, T.M.; Al-Nabulsi, A.A.; Shah, N.P.; Holley, R. Exopolysaccharides as Antimicrobial Agents: Mechanism and Spectrum of Activity. Front. Microbiol. 2021, 12, 1182. [Google Scholar] [CrossRef]
- Sivasankar, P.; Seedevi, P.; Poongodi, S.; Sivakumar, M.; Murugan, T.; Sivakumar, L.; Sivakumar, K.; Balasubramanian, T. Characterization, antimicrobial and antioxidant property of exopolysaccharide mediated silver nanoparticles synthesized by Streptomyces violaceus MM72. Carbohydr. Polym. 2018, 181, 752–759. [Google Scholar] [CrossRef]
- Worsztynowicz, P.; Schmidt, A.O.; Białas, W.; Grajek, W. Identification and partial characterization of proteolytic activity of Enterococcus faecalis relevant to their application in dairy industry. Acta Biochim. Pol. 2019, 66, 61–69. [Google Scholar] [CrossRef]
- Liu, M.; Bayjanov, J.R.; Renckens, B.; Nauta, A.; Siezen, R.J. The proteolytic system of lactic acid bacteria revisited: A genomic comparison. BMC Genom. 2010, 11, 36. [Google Scholar] [CrossRef] [Green Version]
- Nami, Y.; Bakhshayesh, R.V.; Jalaly, H.M.; Lotfi, H.; Eslami, S.; Hejazi, M.A. Probiotic properties of enterococcus isolated from artisanal dairy products. Front. Microbiol. 2019, 10, 300. [Google Scholar] [CrossRef]
- Petrin, S.; Patuzzi, I.; Di Cesare, A.; Tiengo, A.; Sette, G.; Biancotto, G.; Corno, G.; Drigo, M.; Losasso, C.; Cibin, V. Evaluation and quantification of antimicrobial residues and antimicrobial resistance genes in two Italian swine farms. Environ. Pollut. 2019, 255, 113183. [Google Scholar] [CrossRef]
- Ripatti, S.; Tikkanen, E.; Orho-Melander, M.; Havulinna, A.S.; Silander, K.; Sharma, A.; Guiducci, C.; Perola, M.; Jula, A.; Sinisalo, J.; et al. A multilocus genetic risk score for coronary heart disease: Case-control and prospective cohort analyses. Lancet 2010, 376, 1393–1400. [Google Scholar] [CrossRef] [Green Version]
- Zirakzadeh, A.; Patel, R. Vancomycin-resistant enterococci: Colonization, infection, detection, and treatment. Mayo Clin. Proc. 2006, 81, 529–536. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Santella, B.; Folliero, V.; Della Rocca, M.T.; Zannella, C.; Pignataro, D.; Greco, G.; Montella, F.; Folgore, A.; Galdiero, M.; Galdiero, M.; et al. Distribution of antibiotic resistance among Enterococcus spp. isolated from 2017 to 2018 at the University Hospital. Int. J. Mol. Clin. Microbiol. 2019, 9, 1197–1204. [Google Scholar]
- Ayeni, F.A.; Odumosu, B.T.; Oluseyi, A.E.; Ruppitsch, W. Identification and prevalence of tetracycline resistance in enterococci isolated from poultry in Ilishan, Ogun State, Nigeria. J. Pharm. Bioallied Sci. 2016, 8, 69–73. [Google Scholar] [CrossRef]
- Chopra, I.; Roberts, M. Tetracycline Antibiotics: Mode of Action, Applications, Molecular Biology, and Epidemiology of Bacterial Resistance. Microbiol. Mol. Biol. Rev. 2001, 65, 232–260. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robredo, B.; Torres, C.; Singh, K.V.; Murray, B.E. Molecular analysis of Tn1546 in vanA-containing Enterococcus spp. isolated from humans and poultry. Antimicrob. Agents Chemother. 2000, 44, 2588–2589. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simjee, S.; White, D.G.; McDermott, P.F.; Wagner, D.D.; Zervos, M.J.; Donabedian, S.M.; English, L.L.; Hayes, J.R.; Walker, R.D. Characterization of Tn1546 in vancomycin-resistant Enterococcus faecium isolated from canine urinary tract infections: Evidence of gene exchange between human and animal enterococci. J. Clin. Microbiol. 2002, 40, 4659–4665. [Google Scholar] [CrossRef] [Green Version]
- Ranotkar, S.; Kumar, P.; Zutshi, S.; Prashanth, K.S.; Bezbaruah, B.; Anand, J.; Lahkar, M. Vancomycin-resistant enterococci: Troublemaker of the 21st century. J. Glob. Antimicrob. Resist. 2014, 2, 205–212. [Google Scholar] [CrossRef] [PubMed]
- Thumu, S.C.R.; Halami, P.M. Acquired Resistance to Macrolide-Lincosamide-Streptogramin Antibiotics in Lactic Acid Bacteria of Food Origin. Indian J. Microbiol. 2012, 52, 530–537. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Antibiotics in Animal Farming|Compassion in World Farming. Available online: https://www.ciwf.org.uk/research/food-and-human-health/antibiotics-in-animal-farming/ (accessed on 27 July 2021).
- Terzić-Vidojević, A.; Veljović, K.; Begović, J.; Filipić, B.; Popović, D.; Tolinački, M.; Miljković, M.; Kojić, M.; Golić, N. Diversity and antibiotic susceptibility of autochthonous dairy enterococci isolates: Are they safe candidates for autochthonous starter cultures? Front. Microbiol. 2015, 6, 954. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jahan, M.; Zhanel, G.G.; Sparling, R.; Holley, R.A. Horizontal transfer of antibiotic resistance from Enterococcus faecium of fermented meat origin to clinical isolates of E. faecium and Enterococcus faecalis. Int. J. Food Microbiol. 2015, 199, 78–85. [Google Scholar] [CrossRef]
- Tran, T.H.T.; Everaert, N.; Bindelle, J. Review on the effects of potential prebiotics on controlling intestinal enteropathogens Salmonella and Escherichia coli in pig production. J. Anim. Physiol. Anim. Nutr. 2018, 102, 17–32. [Google Scholar] [CrossRef] [Green Version]
- Herrero, I.A.; Fernández-Garayzábal, J.F.; Moreno, M.A.; Domínguez, L. Dogs Should Be Included in Surveillance Programs for Vancomycin-Resistant Enterococci. J. Clin. Microbiol. 2004, 42, 1384–1385. [Google Scholar] [CrossRef] [Green Version]
- Manson, J.M.; Keis, S.; Smith, J.M.B.; Cook, G.M. Characterization of a vancomycin-resistant Enterococcus faecalis (VREF) isolate from a dog with mastitis: Further evidence of a clonal lineage of VREF in New Zealand. J. Clin. Microbiol. 2003, 41, 3331–3333. [Google Scholar] [CrossRef] [Green Version]
- Peters, J.; Mac, K.; Wichmann-Schauer, H.; Klein, G.; Ellerbroek, L. Species distribution and antibiotic resistance patterns of enterococci isolated from food of animal origin in Germany. Int. J. Food Microbiol. 2003, 88, 311–314. [Google Scholar] [CrossRef]
- Dubin, K.; Pamer, E.G. Enterococci and Their Interactions with the Intestinal Microbiome. Microbiol. Spectr. 2017, 5. [Google Scholar] [CrossRef] [Green Version]

| Advantages of Enterococci | Reference |
| Commensals | |
| Immune homeostasis | [9,10] |
| Immunomodulatory effect | [10,44,45,46,63,64,65,78] |
| Producing bacteriocins against pathogens | [53,73,74,158] |
| Metabolism of carbohydrates and proteins—role in digestion | [14,15] |
| Blocking the spread of putrefactive bacteria | [14] |
| Lowering cholesterol levels | [63,66,75] |
| Protective role against cancer | [76] |
| Probiotics | |
| Biotherapeutic—e.g., chronic sinusitis, bronchitis | [13,50] |
| Bio-preservatives and hygiene indicator in food production | [12,14,15] |
| Dietary supplementation for animals | [46,48,67,69,70,77,78] |
| Starter cultures in dairy products | [11,14,15,80] |
| After treatment with antibiotics and as treatment for vancomycin-resistant enterococci colonization | [178] |
| Disadvantages of Enterococci | Reference |
| Potential pathogens (e.g., urinary tract infections, endocarditis) | [83,84,85,92,96,101,102,103,104,133,134] |
| translocation in the circulatory system (sepsis, bacteremia) | [41,44,94,95,96,97] |
| Nosocomial infection/hospital outbreak | [17,84,86,89,90] |
| Virulence and resistance factors can be transmitted between species or genera by horizontal gene transfer—a problem in hospital settings | [116,147,148,150,164,177] |
| Responsible for allergic reactions | [15] |
| Food spoilage | [15,115,119] |
| Food poisoning (foodborne pathogens) | [15] |
| Polyp formation and colorectal cancer | [18,108,110,111] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Krawczyk, B.; Wityk, P.; Gałęcka, M.; Michalik, M. The Many Faces of Enterococcus spp.—Commensal, Probiotic and Opportunistic Pathogen. Microorganisms 2021, 9, 1900. https://doi.org/10.3390/microorganisms9091900
Krawczyk B, Wityk P, Gałęcka M, Michalik M. The Many Faces of Enterococcus spp.—Commensal, Probiotic and Opportunistic Pathogen. Microorganisms. 2021; 9(9):1900. https://doi.org/10.3390/microorganisms9091900
Chicago/Turabian StyleKrawczyk, Beata, Paweł Wityk, Mirosława Gałęcka, and Michał Michalik. 2021. "The Many Faces of Enterococcus spp.—Commensal, Probiotic and Opportunistic Pathogen" Microorganisms 9, no. 9: 1900. https://doi.org/10.3390/microorganisms9091900
APA StyleKrawczyk, B., Wityk, P., Gałęcka, M., & Michalik, M. (2021). The Many Faces of Enterococcus spp.—Commensal, Probiotic and Opportunistic Pathogen. Microorganisms, 9(9), 1900. https://doi.org/10.3390/microorganisms9091900