Microbial Community and Short-Chain Fatty Acid Mapping in the Intestinal Tract of Quail
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethical Statement
2.2. Animals, Experimental Design and Sample Collection
2.3. DNA Extraction, Amplification and Sequencing
2.4. Analysis of Short-Chain Fatty Acids
2.5. qPCR Analyses of Key Bacteria and Genes in Butyrate Production
2.6. Statistics
3. Results
3.1. Microbial Complexity
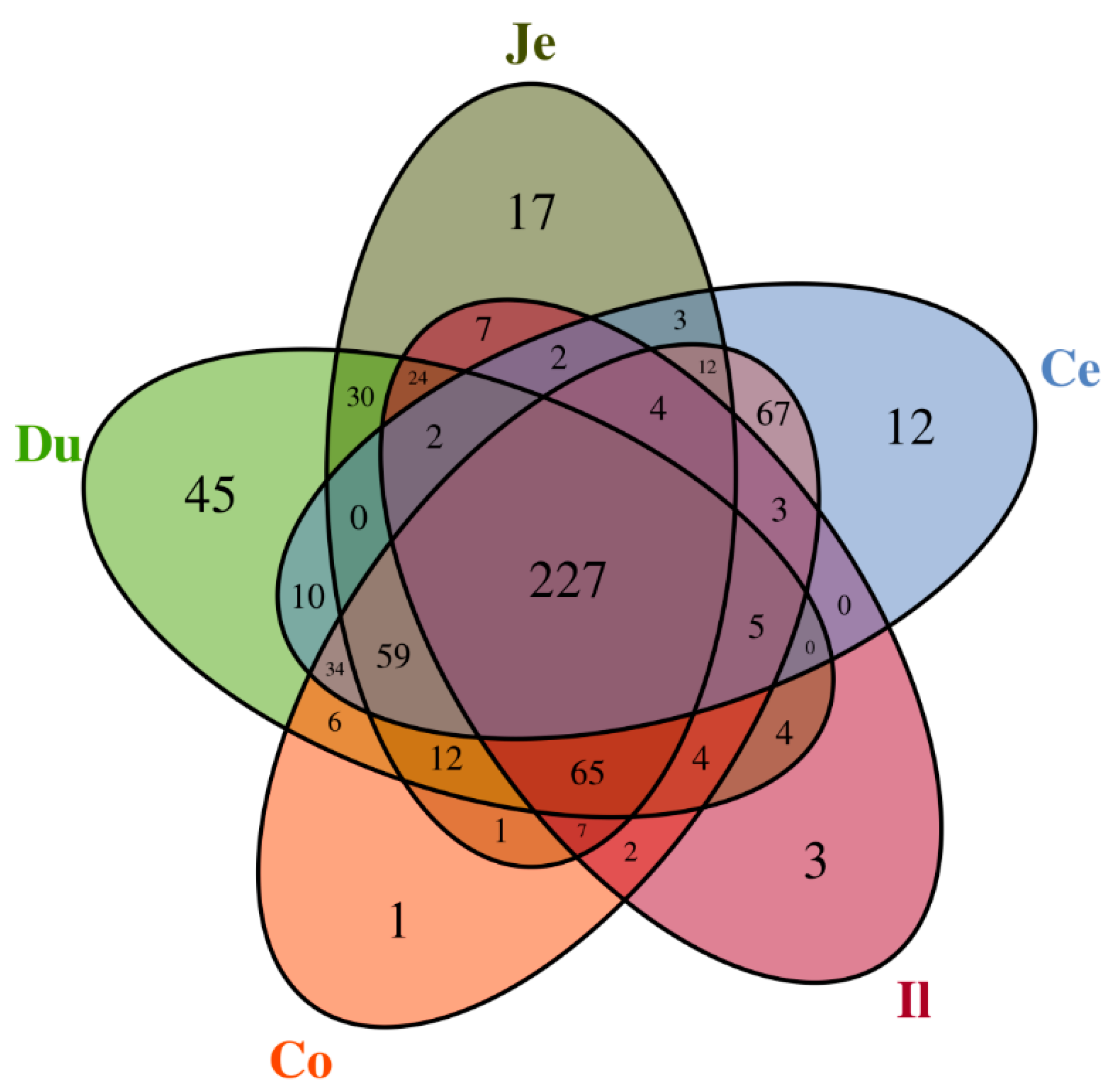
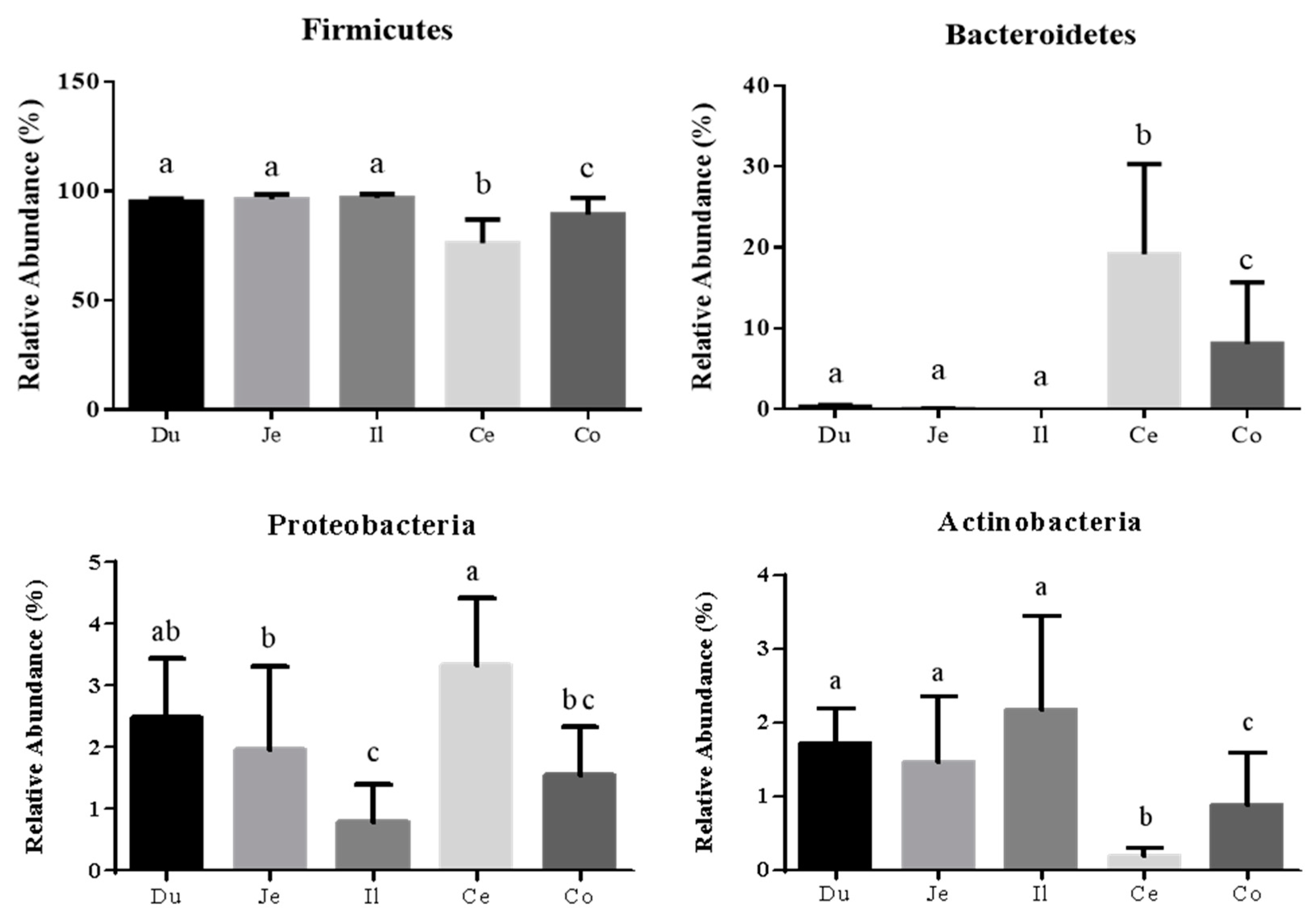
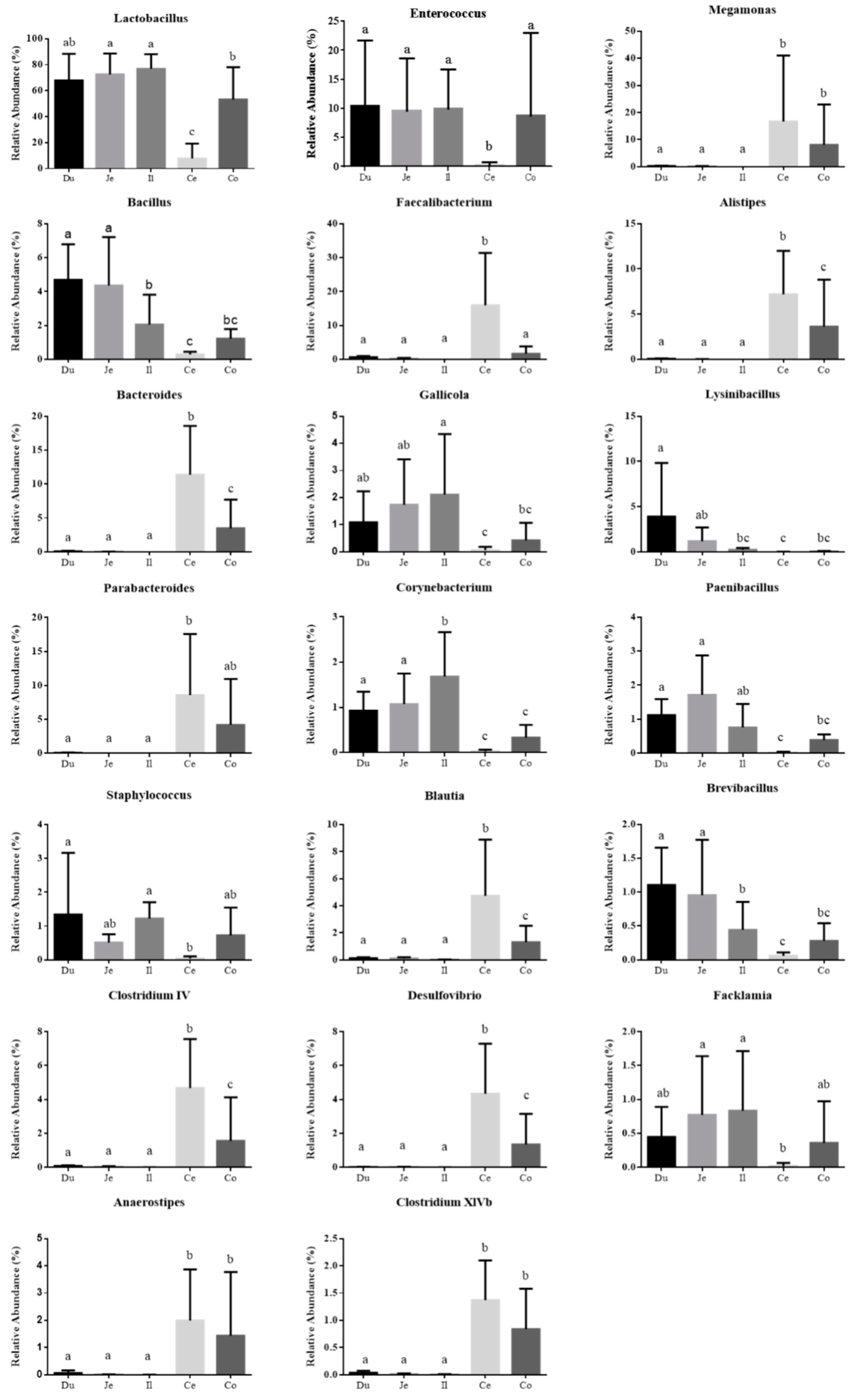
3.2. Microbial Community Composition across the Intestinal Tract
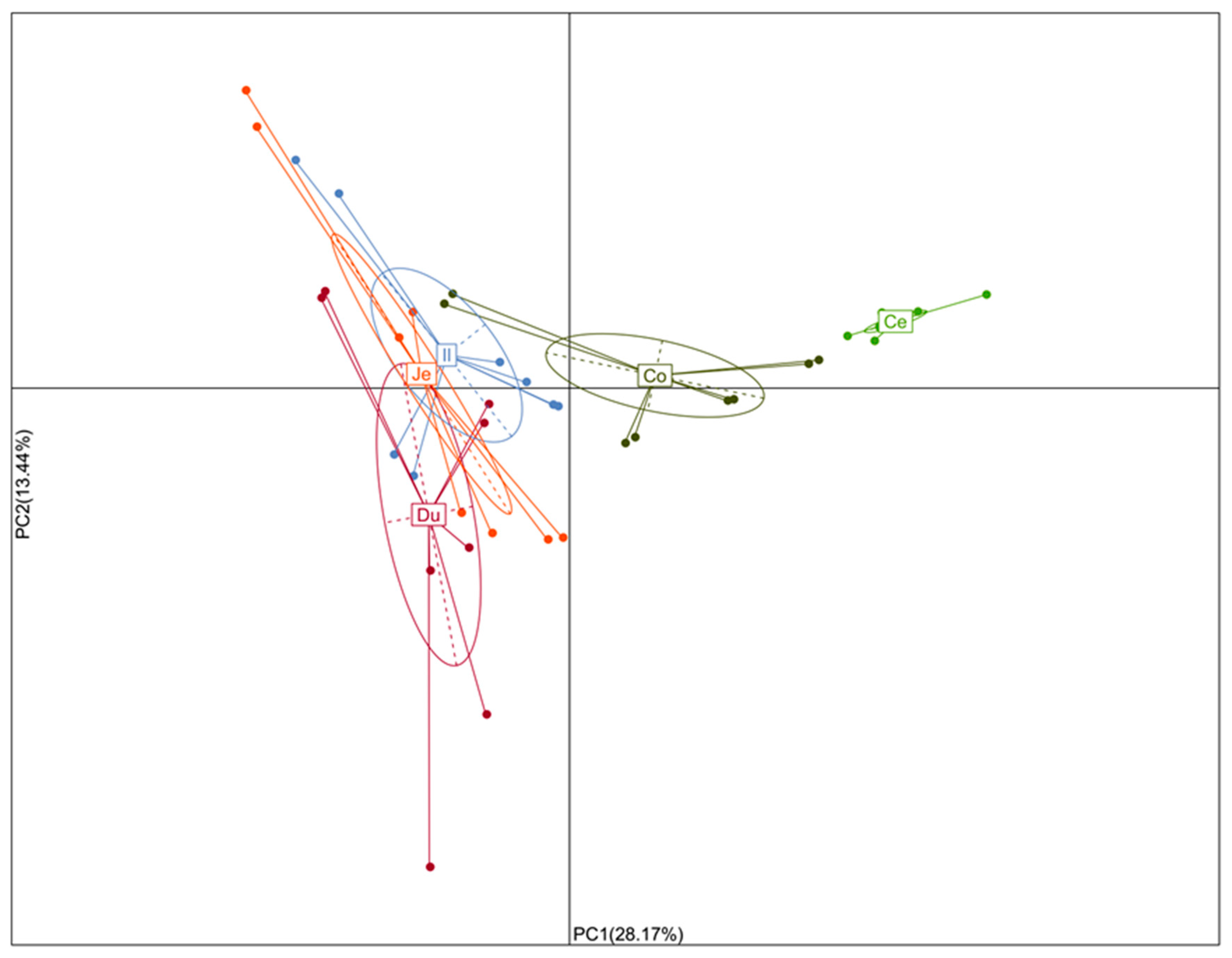
3.3. Similarity of Microbiota across the Intestinal Tract
3.4. SCFAs in Different Intestinal Tract Sections and Its Correlation with Microbiota
3.5. Genes for Butyrate Synthesis in Different Intestinal Tract Sections
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Cullere, M.; Woods, M.J.; van Emmenes, L.; Pieterse, E.; Hoffman, L.C.; Dalle Zotte, A. Hermetia illucens Larvae Reared on Different Substrates in Broiler Quail Diets: Effect on Physicochemical and Sensory Quality of the Quail Meat. Animals 2019, 9, 525. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arantes da Silva, W.; Naiverti Elias, A.H.; Aricetti, J.A.; Sakamoto, M.I.; Murakami, A.E.; Marques Gomes, S.T.; Visentainer, J.V.; Evelázio de Souza, N.; Matsushita, M. Quail egg yolk (Cotumix cotttmix japonica) enriched with omega-3 fatty acids. LWT-Food Sci. Technol. 2009, 42, 660–663. [Google Scholar] [CrossRef]
- Padgett, C.A.; Ivey, W.D. Coturnix quail as a laboratory research animal. Science 1959, 129, 267–268. [Google Scholar] [CrossRef] [PubMed]
- Huss, D.; Poynter, G.; Lansford, R. Japanese quail (Coturnix japonica) as a laboratory animal model. Lab Anim. (NY) 2008, 37, 513–519. [Google Scholar] [CrossRef]
- Ruuskanen, S.; Rainio, M.; Kuosmanen, V.; Laihonen, M.; Saikkonen, K.; Saloniemi, I.; Helander, M. Female preference and adverse developmental effects of glyphosate-based herbicides on ecologically relevant traits in Japanese quails. Environ. Sci. Technol. 2020, 54, 1128–1135. [Google Scholar] [CrossRef]
- Wilkinson, N.; Hughes, R.J.; Aspden, W.J.; Chapman, J.; Moore, R.J.; Stanley, D. The gastrointestinal tract microbiota of the Japanese quail, Coturnix japonica. Appl. Microbiol. Biotechnol. 2016, 100, 4201–4209. [Google Scholar] [CrossRef] [PubMed]
- Mackie, R.I.; Sghir, A.; Gaskins, H.R. Developmental microbial ecology of the neonatal gastrointestinal tract. Am. J. Clin. Nutr. 1999, 69, 1035S–1045S. [Google Scholar] [CrossRef]
- Zoetendal, E.G.; Collier, C.T.; Koike, S.; Mackie, R.I.; Gaskins, H.R. Molecular ecological analysis of the gastrointestinal microbiota: A review. J. Nutr. 2004, 134, 465–472. [Google Scholar] [CrossRef]
- Musso, G.; Gambino, R.; Cassader, M. Interactions between gut microbiota and host metabolism predisposing to obesity and diabetes. Annu. Rev. Med. 2011, 62, 361–380. [Google Scholar] [CrossRef]
- Kamada, N.; Seo, S.U.; Chen, G.Y.; Núñez, G. Role of the gut microbiota in immunity and inflammatory disease. Nat. Rev. Immunol. 2013, 13, 321–335. [Google Scholar] [CrossRef]
- Pan, D.; Yu, Z. Intestinal microbiome of poultry and its interaction with host and diet. Gut Microbes 2014, 5, 108–119. [Google Scholar] [CrossRef] [PubMed]
- Koh, A.; De Vadder, F.; Kovatcheva-Datchary, P.; Bäckhed, F. From dietary fiber to host physiology: Short-chain fatty acids as key bacterial metabolites. Cell 2016, 165, 1332–1345. [Google Scholar] [CrossRef] [Green Version]
- Joseph, J.; Depp, C.; Shih, P.B.; Cadenhead, K.S.; Schmid-Schönbein, G. Modified mediterranean diet for enrichment of short chain fatty acids: Potential adjunctive therapeutic to target immune and metabolic dysfunction in schizophrenia? Front. Neurosci. 2017, 11, 155. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ohira, H.; Tsutsui, W.; Fujioka, Y. Are short chain fatty acids in gut microbiota defensive players for inflammation and atherosclerosis? J. Atheroscler. Thromb. 2017, 24, 660–672. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Su, H.; McKelvey, J.; Rollins, D.; Zhang, M.; Brightsmith, D.J.; Derr, J.; Zhang, S. Cultivable bacterial microbiota of northern bobwhite (Colinus virginianus): A new reservoir of antimicrobial resistance? PLoS ONE 2014, 9, e99826. [Google Scholar] [CrossRef] [PubMed]
- Oakley, B.B.; Lillehoj, H.S.; Kogut, M.H.; Kim, W.K.; Maurer, J.J.; Pedroso, A.; Lee, M.D.; Collett, S.R.; Johnson, T.J.; Cox, N.A. The chicken gastrointestinal microbiome. FEMS Microbiol. Lett. 2014, 360, 100–112. [Google Scholar] [CrossRef] [PubMed]
- Robert, C.E.; Haas, B.J.; Clemente, J.C.; Quince, C.; Knight, R. UCHIME improve sensitivity and speed of chimera detection. Bioinformatics 2011, 27, 2194–2200. [Google Scholar]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Pena, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [Green Version]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef] [Green Version]
- Xiao, Y.; Li, K.; Xiang, Y.; Zhou, W.; Gui, G.; Yang, H. The fecal microbiota composition of boar Duroc, Yorkshire, Landrace and Hampshire pigs. Asian-Australas. J. Anim. Sci. 2017, 30, 1456–1463. [Google Scholar] [CrossRef] [Green Version]
- Xu, J.; Verbrugghe, A.; Lourenço, M.; Cools, A.; Liu, D.J.X.; Van de Wiele, T.; Marzorati, M.; Eeckhaut, V.; Van Immerseel, F.; Vanhaecke, L.; et al. The response of canine faecal microbiota to increased dietary protein is influenced by body condition. BMC. Vet. Res. 2017, 13, 374. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vasaï, F.; Ricaud, K.B.; Cauquil, L.; Daniel, P.; Peillod, C.; Gontier, K.; Tizaoui, A.; Bouchez, O.; Combes, S.; Davail, S. Lactobacillus sakei modulates mule duck microbiota in ileum and ceca during overfeeding. Poult. Sci. 2014, 93, 916–925. [Google Scholar] [CrossRef] [PubMed]
- Huang, P.; Zhang, Y.; Xiao, K.; Jiang, F.; Wang, H.; Tang, D.; Liu, D.; Liu, B.; Liu, Y.; He, X.; et al. The chicken gut metagenome and the modulatory effects of plant-derived benzylisoquinoline alkaloids. Microbiome 2018, 6, 211. [Google Scholar] [CrossRef] [Green Version]
- Zhao, L.; Zhang, F.; Ding, X.; Wu, G.; Lam, Y.Y.; Wang, X.; Fu, H.; Xue, X.; Lu, C.; Ma, J.; et al. Gut bacteria selectively promoted by dietary fibers alleviate type 2 diabetes. Science 2018, 359, 1151–1156. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sommer, F.; Bäckhed, F. The gut microbiota--masters of host development and physiology. Nat. Rev. Microbiol. 2013, 11, 227–238. [Google Scholar] [CrossRef]
- Lee, W.J.; Hase, K. Gut microbiota-generated metabolites in animal health and disease. Nat. Chem. Biol. 2014, 10, 416–424. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Zhai, S.; Xia, Y.; Wang, H.; Ruan, D.; Zhou, T.; Zhu, Y.; Zhang, H.; Zhang, M.; Ye, H.; et al. Ochratoxin A induces liver inflammation: Involvement of intestinal microbiota. Microbiome 2019, 7, 151. [Google Scholar] [CrossRef]
- Corrigan, A.; de Leeuw, M.; Penaud-Frézet, S.; Dimova, D.; Murphy, R.A. Phylogenetic and functional alterations in bacterial community compositions in broiler ceca as a result of mannan oligosaccharide supplementation. Appl. Environ. Microbiol. 2015, 81, 3460–3470. [Google Scholar] [CrossRef] [Green Version]
- Xiao, Y.; Xiang, Y.; Zhou, W.; Chen, J.; Li, K.; Yang, H. Microbial community mapping in intestinal tract of broiler chicken. Poult. Sci. 2017, 96, 1387–1393. [Google Scholar] [CrossRef]
- Wang, S.; Chen, L.; He, M.; Shen, J.; Li, G.; Tao, Z.; Wu, R.; Lu, L. Different rearing conditions alter gut microbiota composition and host physiology in Shaoxing ducks. Sci. Rep. 2018, 8, 7387. [Google Scholar] [CrossRef]
- Yang, H.; Xiao, Y.; Gui, G.; Li, J.; Wang, J.; Li, D. Microbial community and short-chain fatty acid profile in gastrointestinal tract of goose. Poult. Sci. 2018, 97, 1420–1428. [Google Scholar] [CrossRef] [PubMed]
- Waite, D.W.; Taylor, M.W. Exploring the avian gut microbiota: Current trends and future directions. Front. Microbiol. 2015, 6, 673. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Wu, W.; Lee, Y.K.; Xie, J.; Zhang, H. Spatial heterogeneity and co-occurrence of mucosal and luminal microbiome across swine intestinal tract. Front. Microbiol. 2018, 9, 48. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- D’Andreano, S.; Sànchez Bonastre, A.; Francino, O.; Cuscó Martí, A.; Lecchi, C.; Grilli, G.; Giovanardi, D.; Ceciliani, F. Gastrointestinal microbial population of turkey (Meleagris gallopavo) affected by hemorrhagic enteritis virus. Poult. Sci. 2017, 96, 3550–3558. [Google Scholar] [CrossRef] [PubMed]
- Mead, G.C. Microbes of the avian cecum: Types present and substrates utilized. J. Exp. Zool. Suppl. 1989, 3, 48–54. [Google Scholar] [CrossRef] [PubMed]
- Albenberg, L.; Esipova, T.V.; Judge, C.P.; Bittinger, K.; Chen, J.; Laughlin, A.; Grunberg, S.; Baldassano, R.N.; Lewis, J.D.; Li, H.; et al. Correlation between intraluminal oxygen gradient and radial partitioning of intestinal microbiota. Gastroenterology 2014, 147, 1055–1063. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wexler, A.G.; Goodman, A.L. An insider’s perspective: Bacteroides as a window into the microbiome. Nat. Microbiol. 2017, 2, 17026. [Google Scholar] [CrossRef] [Green Version]
- Yang, H.; Xiao, Y.; Wang, J.; Xiang, Y.; Gong, Y.; Wen, X.; Li, D. Core gut microbiota in Jinhua pigs and its correlation with strain, farm and weaning age. J. Microbiol. 2018, 56, 346–355. [Google Scholar] [CrossRef]
- Zhang, J.; Guo, Z.; Xue, Z.; Sun, Z.; Zhang, M.; Wang, L.; Wang, G.; Wang, F.; Xu, J.; Cao, H.; et al. A phylo-functional core of gut microbiota in healthy young Chinese cohorts across lifestyles, geography and ethnicities. ISME J. 2015, 9, 1979–1990. [Google Scholar] [CrossRef]
- Louis, P.; Duncan, S.H.; McCrae, S.I.; Millar, J.; Jackson, M.S.; Flint, H.J. Restricted distribution of the butyrate kinase pathway among butyrate-producing bacteria from the human colon. J. Bacteriol. 2004, 186, 2099–2106. [Google Scholar] [CrossRef] [Green Version]
- Vital, M.; Penton, C.R.; Wang, Q.; Young, V.B.; Antonopoulos, D.A.; Sogin, M.L.; Morrison, H.G.; Raffals, L.; Chang, E.B.; Huffnagle, G.B.; et al. A gene-targeted approach to investigate the intestinal butyrate-producing bacterial community. Microbiome 2013, 1, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| Item | Starter Feed | Grower Feed |
|---|---|---|
| Ingredient (% of diet) | ||
| Corn | 53.24 | 58.88 |
| Soybean meal | 38.66 | 32.97 |
| Soybean oil | 2.80 | 3.29 |
| CaHPO4 | 1.26 | 1.09 |
| Limestone | 1.31 | 1.41 |
| NaCl | 1.26 | 1.09 |
| Met | 0.24 | 0.22 |
| Lys | 0.03 | 0.05 |
| Premix | 1.00 | 1.00 |
| Nutrient level | ||
| Metabolizable energy, MJ/kg | 12.13 | 12.65 |
| Crude protein, % | 20.80 | 19.50 |
| Lys | 1.28 | 1.19 |
| Met | 0.52 | 0.45 |
| Calcium, % | 0.91 | 0.85 |
| Phosphorus, % | 0.57 | 0.53 |
| Item | Primers (5′–3′) | Reference |
|---|---|---|
| Total Bacteria | fwd CGGYCCAGACTCCTACGGG rev TTACCGCGGCTGCTGGCA | [21] |
| Firmicutes | fwd GGAGYATGTGGTTTAATTCGAAGCA rev AGCTGACGACAACCATGCAC | [21] |
| Bacteroidetes | fwd GGARCATGTGGTTTAATTCGATGAT rev AGCTGACGACAACCATGCAG | [21] |
| Bacteroides | fwd GAGAGGAAGGTCCCCCAC rev CGCTACTTGGCTGGTTCAG | [22] |
| Butyryl-CoA:acetate-CoA transferase | fwd AAGGATCTCGGIRTICAYWSIGARATG rev GAGGTCGTCICKRAAITYIGGRTGNGC | [21] |
| Butyrate kinase | fwd TGCTGTWGTTGGWAGAGGYGGA rev GCAACIGCYTTTTGATTTAATGCATGG | [21] |
| Item | Duodenum | Jejunum | Ileum | Cecum | Colorectum |
|---|---|---|---|---|---|
| Sequencing Number | 60280 ± 3003 | 60591 ± 2437 | 61195 ± 2438 | 60101 ± 3778 | 59540 ± 1336 |
| OTUs | 259 ± 23 | 237 ± 68 | 201 ± 40 | 275 ± 39 | 270 ± 45 |
| Chao1 | 281.67 ± 29.79 ab | 271.10 ± 75.05 ab | 257.66 ± 29.92 b | 310.72 ± 51.10 a | 322.06 ± 61.66 a |
| Shannon | 3.07 ± 0.45 bc | 2.78 ± 0.77 c | 2.61 ± 0.47 c | 4.83 ± 0.52 a | 3.72 ± 0.76 ab |
| Simpson | 0.71 ± 0.08 b | 0.68 ± 0.10 b | 0.68 ± 0.07 b | 0.89 ± 0.06 a | 0.81 ± 0.08 ab |
| Good’s coverage | 0.9992 ± 0.0002 | 0.9990 ± 0.0003 | 0.9989 ± 0.0000 | 0.9991 ± 0.0002 | 0.9989 ± 0.0003 |
| Item | Duodenum | Jejunum | Ileum | Cecum | Colorectum |
|---|---|---|---|---|---|
| Total Bacteria | 5.98 ± 1.07 a | 7.11 ± 0.40 bc | 8.83 ± 0.68 c | 10.97 ± 1.31 d | 11.05 ± 0.64 d |
| Firmicutes | 5.93 ± 0.68 a | 7.02 ± 0.75 b | 7.93 ± 0.81 b | 10.06 ± 1.42 c | 10.88 ± 0.94 c |
| Bacteroidetes | 4.76 ± 0.51 a | 5.08 ± 0.82 ab | 6.45 ± 0.36 b | 9.63 ± 0.72 c | 9.46 ± 0.39 c |
| Bacteroides | 4.80 ± 0.63 a | 5.05 ± 0.42 b | 5.87 ± 0.51 b | 9.02 ± 0.53 c | 8.77 ± 0.45 c |
| Item | Duodenum | Jejunum | Ileum | Cecum | Colorectum |
|---|---|---|---|---|---|
| Acetate | 0.0415 ± 0.0361 a | 0.0477 ± 0.0243 a | 0.0866 ± 0.0764 b | 1.3092 ± 0.4760 c | 0.7251 ± 0.1653 d |
| Propionate | 0.0303 ± 0.0218 a | 0.0287 ± 0.0267 a | 0.0401 ± 0.0423 a | 0.6788 ± 0.2488 b | 0.3898 ± 0.1131 c |
| Butyrate | — | — | 0.0261 ± 0.0236 | 0.5482 ± 0.1595 | 0.2348 ± 0.1243 |
| Isobutyrate | — | — | — | 0.0511 ± 0.0044 | 0.0440 ± 0.0074 |
| Valerate | — | — | — | 0.0319 ± 0.0251 | 0.0232 ± 0.0315 |
| Isovalerate | — | — | — | 0.0265 ± 0.0353 | — |
| Item | Duodenum | Jejunum | Ileum | Cecum | Colorectum |
|---|---|---|---|---|---|
| Butyryl-CoA:acetate-CoA transferase | 4.02 ± 0.37 a | 3.73 ± 0.50 a | 3.99 ± 0.62 a | 6.70 ± 0.27 b | 6.03 ± 0.44 c |
| Butyrate kinase | 4.13 ± 0.29 a | 4.21 ± 0.51 a | 5.41 ± 0.59 b | 6.82 ± 0.51 c | 5.68 ± 0.49 d |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Du, X.; Xiang, Y.; Lou, F.; Tu, P.; Zhang, X.; Hu, X.; Lyu, W.; Xiao, Y. Microbial Community and Short-Chain Fatty Acid Mapping in the Intestinal Tract of Quail. Animals 2020, 10, 1006. https://doi.org/10.3390/ani10061006
Du X, Xiang Y, Lou F, Tu P, Zhang X, Hu X, Lyu W, Xiao Y. Microbial Community and Short-Chain Fatty Acid Mapping in the Intestinal Tract of Quail. Animals. 2020; 10(6):1006. https://doi.org/10.3390/ani10061006
Chicago/Turabian StyleDu, Xizhong, Yun Xiang, Fangfang Lou, Pingguang Tu, Xiaojun Zhang, Xujin Hu, Wentao Lyu, and Yingping Xiao. 2020. "Microbial Community and Short-Chain Fatty Acid Mapping in the Intestinal Tract of Quail" Animals 10, no. 6: 1006. https://doi.org/10.3390/ani10061006




