The Study of the Response of Fat Metabolism to Long-Term Energy Stress Based on Serum, Fatty Acid and Transcriptome Profiles in Yaks
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Slaughter Procedure and Sample Collection
2.3. Determination of Fat Content and Fatty Acid Profiles
2.4. Determination of Serum Profiles
2.5. RNA Extraction, Read Mapping and Expression Annotation
2.6. Validation of Differentially Expressed Genes
3. Results
3.1. Fat Content and Fatty Acid Profiles
3.2. Serum Profiles
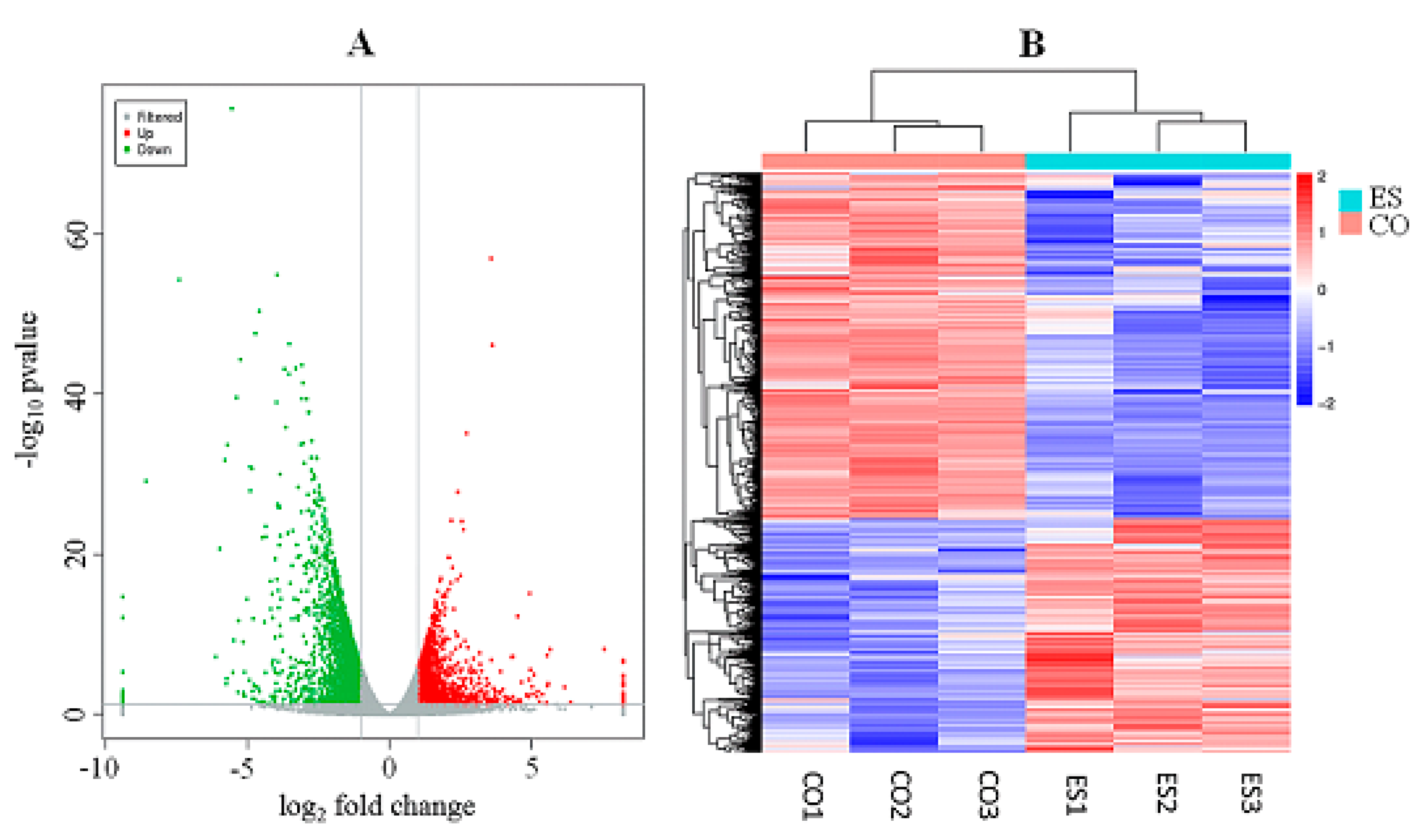
3.3. Identification of Differentially Expressed Genes
3.4. GO and KEGG Analysis of DEGs
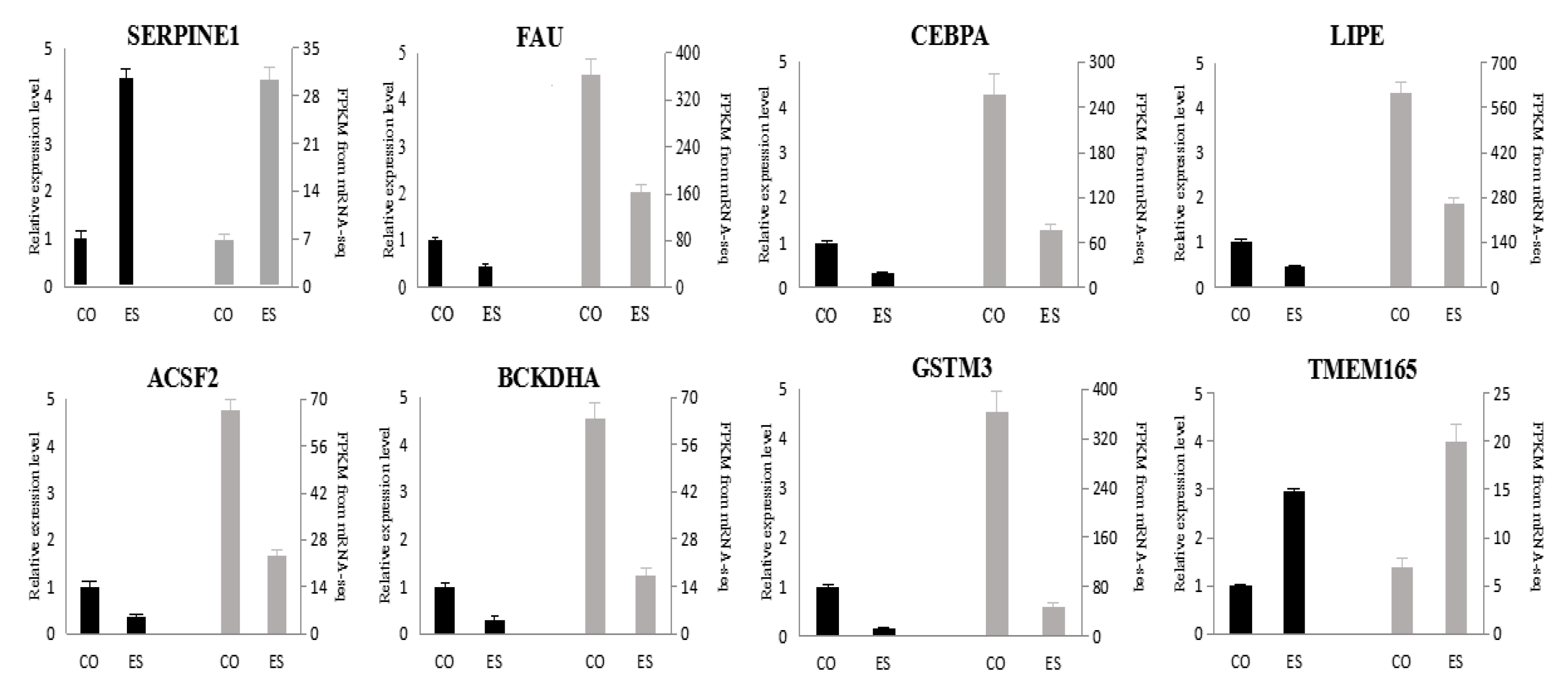
3.5. qPCR Validation
4. Discussion
4.1. The Physiological Status of Yaks under Long-Term ES
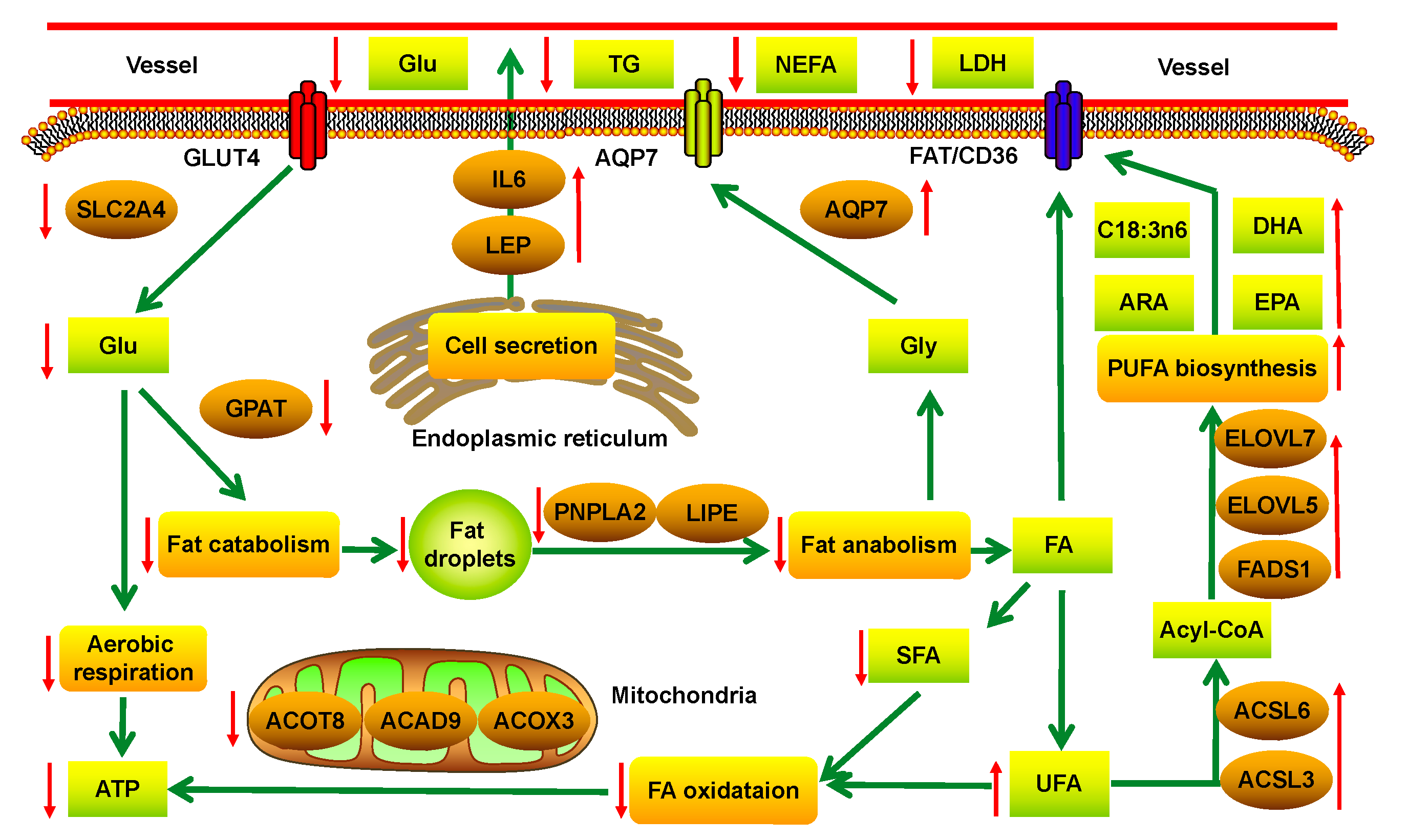
4.2. The Effect of Long-Term ES on Fat Metabolism in Yaks
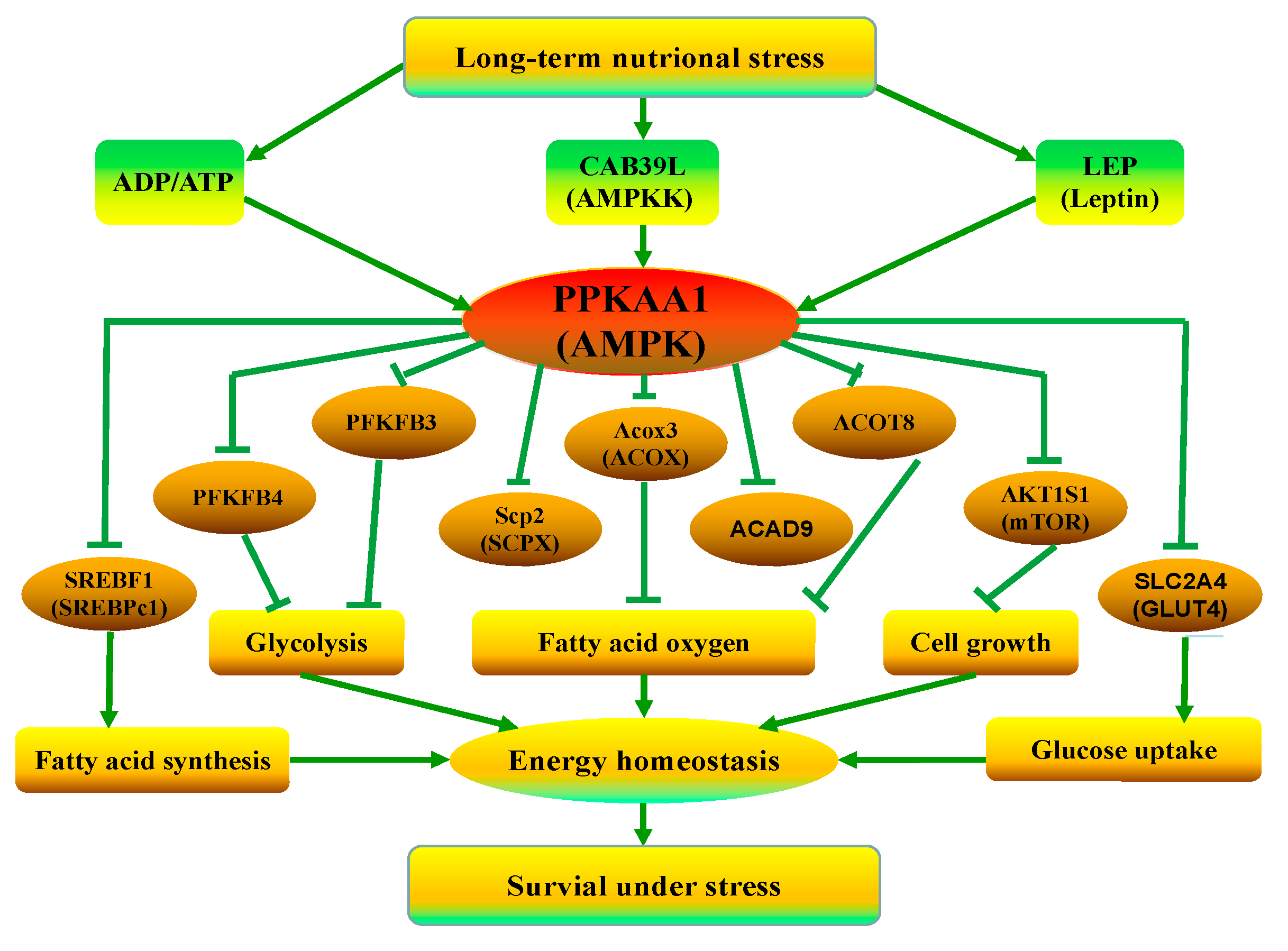
4.3. The Mechanism of the Response of Fat Metabolism to Long-Term ES
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Wen, W.T.; Luo, X.L.; Xia, B.X.; Guan, J.Q.; Nie, Y.Y.; Li, L.; Duan, J.Y.; Suman, S.P.; Sun, Q. Post-mortem oxidative stability of three yak (Bos grunniens) muscles as influenced by animal age. Meat Sci. 2015, 105, 121–125. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.S.; Long, R.J.; kreuzer, M.; Ding, L.M.; Shang, Z.H.; Zhang, Y.; Yang, Y.; Cui, G.X. Importance of functional ingredients in yak milk-derived food on health of Tibetan nomads living under high-altitude stress: A review. Crit. Rev. Food Sci. 2014, 54, 292–302. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.L.; Han, L.; Ma, X.L.; Yu, Q.L.; Zhao, S.N. Effect of mitochondrial apoptotic activation through the mitochondrial membrane permeability transition pore on yak meat tenderness during postmortem aging. Food Chem. 2017, 234, 323–331. [Google Scholar] [CrossRef]
- Hu, R.; Zou, H.W.; Wang, Z.S.; Cao, B.H.; Peng, Q.H.; Jing, X.P.; Wang, Y.X.; Shao, Y.Q.; Pei, Z.X.; Zhang, X.F.; et al. Nutritional interventions improved rumen functions and promoted compensatory growth of growth-retarded yaks as revealed by integrated transcripts and microbiome analyses. Front. Microbiol. 2019, 10, 318. [Google Scholar] [CrossRef] [PubMed]
- Barboza, P.S.; Jorde, D.G. Monitoring responses to variation in food supply for a migratory waterfowl: American Black Duck (Anas rubripes) in winter. J. Comp. Physiol. B 2018, 188, 831–842. [Google Scholar] [CrossRef] [PubMed]
- Masoodi, M.; Kuda, O.; Rossmeisl, M.; Flachs, P.; Kopecky, J. Lipid signaling in adipose tissue: Connecting inflammation & metabolism. Biochim. Biophys. Acta 2015, 1851, 503–518. [Google Scholar]
- Angeli, E.; Trionfinia, V.; Gareisa, N.C.; Matiller, V.; Huber, E.; Reya, F.; Salvettia, N.R.; Ortegaa, H.H.; Hein, G.J. Protein and gene expression of relevant enzymes and nuclear receptor of hepatic lipid metabolism in grazing dairy cattle during the transition period. Res. Vet. Sci. 2019, 123, 223–231. [Google Scholar] [CrossRef]
- Gui, L.S.; Hong, J.Y.; Raza, S.H.A.; Zan, L.S. Genetic variants in SIRT3 transcriptional regulatory region affect promoter activity and fat deposition in three cattle breeds. Mol. Cell. Probes 2017, 32, 40–45. [Google Scholar] [CrossRef]
- Strieder-Barboza, C.; Contreras, G.A. Fetuin-A modulates lipid mobilization in bovine adipose tissue by enhancing lipogenic activity of adipocytes. J. Dairy Sci. 2019, 102, 4628–4638. [Google Scholar] [CrossRef]
- Jiang, R.; Li, H.; Huang, Y.Z.; Lan, X.Y.; Lei, C.Z.; Chen, H. Transcriptome profiling of lncRNA related to fat tissues of Qinchuan cattle. Gene 2020, 742, 144587. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, Y.Q.; Zhang, X.Z.; Wang, D.C.; Jin, G.; Li, B.; Xu, F.; Cheng, J.; Zhang, F.; Wu, S.J.; et al. The comprehensive liver transcriptome of two cattle breeds with different intramuscular fat content. Biochem. Biophys. Res. Commun. 2017, 490, 1018–1025. [Google Scholar] [CrossRef] [PubMed]
- McFadden, J.W. Review: Lipid biology in the periparturient dairy cow: Contemporary perspectives. Animal 2020, 14, 165–175. [Google Scholar] [CrossRef] [PubMed]
- Strömqvist, M.; Olsson, J.A.; Kärrman, A.; Brunström, B. Transcription of genes involved in fat metabolism in chicken embryos exposed to the peroxisome proliferator-activated receptor alpha (PPARα) agonist GW7647 or to perfluorooctane sulfonate (PFOS) or perfluorooctanoic acid (PFOA). Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2012, 156, 29–36. [Google Scholar] [CrossRef]
- Na, W.; Wu, Y.Y.; Gong, P.F.; Wu, C.Y.; Cheng, B.H.; Wang, Y.X.; Wang, N.; Du, Z.Q.; Li, H. Embryonic transcriptome and proteome analyses on hepatic lipid metabolism in chickens divergently selected for abdominal fat content. BMC Genom. 2018, 19, 384. [Google Scholar] [CrossRef]
- Liu, L.; Liu, X.J.; Cui, H.X.; Liu, R.R.; Zhao, G.P.; Wen, J. Transcriptional insights into key genes and pathways controlling muscle lipidmetabolism in broiler chickens. BMC Genom. 2019, 20, 863. [Google Scholar] [CrossRef]
- Sun, L.M.; Ling, Y.H.; Jiang, J.H.; Wang, D.T.; Wang, J.X.; Li, J.Y.; Wang, X.D.; Wang, H.L. Differential mechanisms regarding triclosan vs. bisphenol A and fluorene-9-bisphenol induced zebrafish lipid-metabolism disorders by RNA-Seq. Chemosphere 2020, 251, 126318. [Google Scholar] [CrossRef]
- Sciascia, Q.L.; Daş, G.; Maak, S.; Kalbe, C.; Metzler-Zebeli, B.U.; Metges, C.C. Transcript profile of skeletal muscle lipid metabolism genes affected by diet in a piglet model of low birth weight. PLoS ONE 2019, 14, e0224484. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.Q.; Huang, Z.Y.; Zhao, W.J.; Li, M.X.; Li, C.C. Transcriptome analysis reveals long intergenic non-voding RNAs contributed to intramuscular fat content differences between Yorkshire and Wei pigs. Int. J. Mol. Sci. 2020, 21, 1732. [Google Scholar] [CrossRef]
- Tilley, J.M.A.; Terry, R.A. A two-stage technique for the in vitro digestion of forage crops. Grass Forage Sci. 1963, 18, 104–111. [Google Scholar] [CrossRef]
- Takahashi, T.; Jones, R.; Kemp, D. Steapdy-state modeling for better understanding of current livestock production systems and for exploring optimal short-term strategies.Development of sustainable livestock systems on grasslands in north-western China. ACIAR Proceedings. In Proceedings of the XXI International Grassland Congress and VIII International Rangeland Congress, Hohhot, China, 28 June 2008; pp. 26–35, ISBN 9781921615456/9781921615463. [Google Scholar]
- AOAC. Official Method 991. 36. Fat (Crude) in Meat and Meat Products Solvent Extraction (Submersion) Method. In Official Methods of Analysis; AOAC: Washington, DC, USA, 1996. [Google Scholar]
- Song, S.Z.; Wu, J.P.; Zhao, S.G.; Casper, D.P.; He, B.; Liu, T.; Lang, X.; Gong, X.Y.; Liu, L.S. The effect of energy restriction on fatty acid profiles of longissimus dorsi and tissue adipose depots in sheep. J. Anim. Sci. 2017, 95, 3940–3948. [Google Scholar] [CrossRef]
- Keay, G.; Doxey, D.L. Serum albumin values from healthy cattle, sheep and horses determined by the immediate bromocresol green reaction and by agarose gel electrophoresis. Res. Vet. Sci. 1983, 35, 58–60. [Google Scholar] [CrossRef]
- Xu, C.; Liu, G.W.; Li, X.B.; Xia, C.; Zhang, H.Y.; Wang, Z. Decreased complete oxidation capacity of fatty acid in the liver of ketotic cows. Asian-Australian J. Anim. 2010, 23, 312–317. [Google Scholar] [CrossRef]
- Yu, H.B.; Zhao, Z.H.; Yu, X.Z.; Li, J.Y.; Lu, C.Y.; Yang, R.J. Bovine lipid metabolism related gene GPAM: Molecular characterization, function identification, and association analysis with fat deposition traits. Gene 2017, 609, 9–18. [Google Scholar] [CrossRef] [PubMed]
- Fang, X.B.; Zhao, Z.H.; Jiang, P.; Yu, H.B.; Xiao, H.; Yang, R.J. Identification of the bovine HSL gene expression profiles and its association with fatty acid composition and fat deposition traits. Meat Sci. 2017, 131, 107–118. [Google Scholar] [CrossRef]
- Stelmanska, E.; Szrok, S.; Swierczynski, J. Progesterone-induced down-regulation of hormone sensitive lipase (Lipe) and up-regulation of G0/G1 switch 2 (G0s2) genes expression in inguinal adipose tissue of female rats is reflected by diminished rate of lipolysis. J. Steroid. Biochem. Mol. Biol. 2015, 147, 31–39. [Google Scholar] [CrossRef]
- Kumar, S.; Deb, R.; Singh, U.; Ganguly, I.; Mandal, D.K.; Singh, R.; Sharma, S.; Sengar, G.; Singh, R.; Kumar, M.; et al. SNPs at exonic region of aquaporin-7 (AQP7) gene may affect semen quality parameters among crossbred bulls. J. Genet. 2014, 93, e108–e112. [Google Scholar] [CrossRef]
- Li, H.D.; Xu, S.Z.; Gao, X.; Ren, H.Y. Structure of the Bovine ACAD8 Gene and the Association of Its Polymorphism with the Production Traits. J. Genet. Genom. 2007, 34, 315–320. [Google Scholar] [CrossRef]
- Leipnitz, G.; Mohsen, A.; Karunanidhi, A.; Seminotti, B.; Roginskaya, V.Y.; Markantone, D.M.; Grings, M.; Mihalik, S.J.; Wipf, P.; Houten, B.V.; et al. Evaluation of mitochondrial bioenergetics, dynamics, Endoplasmic reticulummitochondria crosstalk, and reactive oxygen species in fbroblasts from patients with complex I deficiency. Sci. Rep. 2018, 8, 1165. [Google Scholar] [CrossRef]
- Krause, K.; Weiner, J.; Klöting, S.H.D.N.; Rijntjes, E.; Heiker, J.T.; Gebhardt, C.; Köhrle, J.; Führer, D.; Steinhoff, K.; Hesse, S.; et al. The effects of thyroid hormones on gene expression of Acyl-coenzyme A thioesterases in adipose tissue and liver of mice. Eur. Thyroid. J. 2015, 4, 59–66. [Google Scholar] [CrossRef]
- Daza, A.; Rey, A.I.; Menoyo, D.; Bautista, J.M.; Olivares, A.; López-Bote, C.J. Effect of level of feed restriction during growth and/or fattening on fatty acid composition and lipogenic enzyme activity in heavy pigs. Anim. Feed Sci. Tech. 2007, 138, 61–74. [Google Scholar] [CrossRef]
- Garcia, P.T.; Pensel, N.A.; Sancho, A.M.; Latimori, N.J.; Kloster, A.M.; Amigone, M.A.; Casal, J.J. Beef lipids in relation to animal breed and nutrition in Argentina. Meat Sci. 2008, 79, 500–508. [Google Scholar] [CrossRef] [PubMed]
- Dinh, T.T.N.; Blanton, J.R.; Riley, D.G.; Chase, C.C.; Coleman, S.W.; Phillips, W.A.; Thompson, L.D. Intramuscular fat and fatty acid composition of longissimus muscle from divergent pure breeds of cattle. J. Anim. Sci. 2010, 88, 756–766. [Google Scholar] [CrossRef] [PubMed]
- Jo, J.L.; Hwang, J.H.; Kwon, S.G.; Park, D.H.; Kim, T.W.; Kang, D.G.; Yu, G.E.; Kim, I.S.; Ha, J.G.; Kim, C.W. Association between a non-synonymous HSD17B4 single nucleotide polymorphism and meat-quality traits in Berkshire pigs. Genet. Mol. Res. 2016, 15, 27819726. [Google Scholar] [CrossRef]
- Witkowski, A.; Thweatt, J.; Smith, S. Mammalian ACSF3 protein is a malonyl-CoA synthetase that supplies the chain extender units for mitochondrial fatty acid synthesis. J. Biol. Chem. 2011, 286, 33729–33736. [Google Scholar] [CrossRef]
- Ren, E.; Chen, X.; Yu, S.; Xu, J.; Su, Y.; Zhu, W. Transcriptomic and metabolomic responses induced in the livers of growing pigs by a short-term intravenous infusion of sodium butyrate. Animal 2018, 12, 2318–2326. [Google Scholar] [CrossRef]
- Valente, R.S.; Almeida, T.G.D.; Alves, M.F.; Camargo, J.D.; Basso, A.C.; Belaz, K.R.A.; Eberlin, M.N.; Landim-Alvarenga, F.D.C.; Fontes, P.K.; Nogueira, M.F.G.N. Modulation of long-chain Acyl-CoA synthetase on the development, lipid deposit and cryosurvival of in vitro produced bovine embryos. PLoS ONE 2019, 14, e0220731. [Google Scholar] [CrossRef] [PubMed]
- Zhu, B.; Niu, H.; Zhang, W.G.; Wang, Z.Z.; Liang, Y.H.; Guan, L.; Guo, P.; Chen, Y.; Zhang, L.P.; Guo, Y.; et al. Genome wide association study and genomic prediction for fatty acid composition in Chinese Simmental beef cattle using high density SNP array. BMC Genom. 2017, 18, 464. [Google Scholar] [CrossRef]
- Chen, S.; Hu, Z.G.; He, H.; Liu, X.L. Fatty acid elongase 7 is regulated via SP1 and is involved in lipid accumulation in bovine mammary epithelial cells. J. Cell. Physiol. 2018, 233, 4715–4725. [Google Scholar] [CrossRef]
- Han, C.; Vinsky, M.; Aldai, N.; Dugan, M.E.R.; Li, T.A.M.C. Association analyses of DNA polymorphisms in bovine SREBP-1, LXRα, FADS1 genes with fatty acid composition in Canadian commercial crossbred beef steer. Meat Sci. 2013, 93, 429–436. [Google Scholar] [CrossRef]
- Zhang, X.F.; Pang, S.C.; Liu, C.J.; Wang, H.P.; Ye, D.; Zhu, Z.Y.; Sun, Y.H. A novel dietary source of EPA and DHA: Metabolic engineering of an important freshwater species—Common carp by fat1-transgenesis. Mar. Biotechnol. 2019, 21, 171–185. [Google Scholar] [CrossRef]
- Bouafi, H.; Bencheikh, S.; Krami, A.M.; Morjane, I.; Charoute, H.; Rouba, H.; Saile, R.; Benhnini, F.; Barakat, A. Prediction and structural comparison of deleterious coding nonsynonymous single nucleotide polymorphisms (nsSNPs) in human LEP gene associated with obesity. Biomed. Res. Int. 2019, 2019, 1832084. [Google Scholar] [CrossRef] [PubMed]
- Kristensen, K.K.; Leth-Espensen, K.Z.; Mertens, H.D.T.; Birrane, G.; Meiyappan, M.; Olivecrona, G.; Jørgensen, T.J.D.; Young, S.G.; Ploug, M. Unfolding of monomeric lipoprotein lipase by ANGPTL4: Insight into the regulation of plasma triglyceride metabolism. Proc. Natl. Acad. Sci. USA 2020, 117, 4337–4346. [Google Scholar] [CrossRef] [PubMed]
- Qiao, L.P.; Shetty, S.K.; Spitler, K.M.; Wattez, J.S.; Davies, B.S.J.; Shao, J.H. Obesity reduces maternal blood triglyceride concentrations by reducing angiopoietin-like protein 4 expression in mice. Diabetes 2020, 69, 1100–1109. [Google Scholar] [CrossRef]
- Hardie, D.G. New roles for the LKB1? AMPK pathway. Curr. Opin. Cell Biol. 2005, 17, 167–173. [Google Scholar] [CrossRef] [PubMed]
- Sui, G.G.; Xiao, H.B.; Lu, X.Y.; Sun, Z.L. Naringin activates AMPK resulting in altered expression of SREBPs, PCSK9, and LDLR to reduce body weight in obese C57BL/6J mice. J. Agric. Food Chem. 2018, 66, 8983–8990. [Google Scholar] [CrossRef]
- Li, W.L.; Wong, C.C.; Zhang, X.M.; Kang, W.; Nakatsu, G.; Zhao, Q.F.; Chen, H.R.; Go, M.Y.Y.; Chiu, P.W.Y.; Wang, X.H.; et al. CAB39L elicited an anti-Warburg effect via a LKB1-AMPK-PGC1α axis to inhibit gastric tumorigenesis. Oncogene 2018, 37, 6383–6398. [Google Scholar] [CrossRef]
- Vlavcheski, F.; Hartogh, D.J.D.; Giacca, A.; Tsiani, E. Amelioration of high-insulin-induced skeletal muscle cell insulin resistance by resveratrol is linked to activation of AMPK and restoration of GLUT4 translocation. Nutrients 2020, 12, 914. [Google Scholar] [CrossRef]
- Fan, H.Y.; Zhou, Y.Y.; Wen, H.S.; Zhang, X.Y.; Zhang, K.Q.; Qi, X.; Xu, P.; Li, Y. Genome-wide identification and characterization of glucose transporter (glut) genes in spotted sea bass (Lateolabrax maculatus) and their regulated hepatic expression during short-term starvation. Comp. Biochem. Physiol. Part. D Genom. Proteom. 2019, 30, 217–229. [Google Scholar] [CrossRef]
- Obsen, T.; Faergeman, N.J.; Chung, S.; Martinez, K.; Gobern, S.; Loreau, O.; McIntosh, M. Trans-10, cis-12 conjugated linoleic acid decreases de novo lipid synthesis in human adipocytes. J. Nutr. Biochem. 2012, 23, 580–590. [Google Scholar] [CrossRef] [PubMed]
- Cuyàs, E.; Corominas-Faja, B.; Joven, J.; Menendez, J.A. Cell cycle regulation by the nutrient-sensing mammalian target of rapamycin (mTOR) pathway. Methods Mol. Biol. 2014, 1170, 113–144. [Google Scholar] [PubMed]
- Düvel, K.; Yecies, J.; Menon, S.; Raman, P.; Lipovsky, A.; Souza, A.; Triantafellow, E.; Ma, Q.; Gorski, R.; Cleaver, S.; et al. Activation of a metabolic gene regulatory network downstream of mTOR complex 1. Mol. Cell. 2010, 39, 171–183. [Google Scholar] [CrossRef] [PubMed]
- Ozturk, D.G.; Kocak, M.; Akcay, A.; Kinoglu, K.; Kara, E.; Buyuk, Y.; Kazan, H.; Gozuacik, D. MITF-MIR211 axis is a novel autophagy amplifier system during cellular stress. Autophagy 2019, 15, 375–390. [Google Scholar] [CrossRef]
- Tuttle, M.; Dalman, M.R.; Liu, Q.; Londraville, R.L. Leptin-a mediates transcription of genes that participate in central endocrine and phosphatidylinositol signaling pathways in 72-hour embryonic zebrafish (Danio rerio). Peer J. 2019, 7, e6848. [Google Scholar] [CrossRef] [PubMed]
- Yuan, D.W.; Zhou, S.; Liu, S.N.; Li, K.; Zhao, H.G.; Long, S.H.; Liu, H.H.; Xie, Y.F.; Su, Y.L.; Yu, F.W.; et al. The AMPK-PP2A axis in insect fat body is activated by 20-hydroxyecdysone to antagonize insulin/IGF signaling and restrict growth rate. Proc. Natl. Acad. Sci. USA 2020, 117, 9292–9301. [Google Scholar] [CrossRef] [PubMed]
- Strembitska, A.; Mancini, S.J.; Gamwell, J.M.; Palmer, T.M.; Baillie, G.S.; Salt, L.P. A769662 inhibits insulin-stimulated Akt activation in human macrovascular endothelial cells independent of AMP-activated protein kinase. Int. J. Mol. Sci. 2018, 19, 3886. [Google Scholar] [CrossRef] [PubMed]
| Gene Symbol | Forward Primer (5′ -> 3′) | Reverse Primer (5′ -> 3′) | Product Size (bp) |
|---|---|---|---|
| SERPINE1 | CGTCCAGAGAGAGCCACA | ATCCGCATCCTGAATTTCG | 118 |
| FAU | CTACGCACTCTCGAGGTGA | AGGAGCAGGACTTGATCT | 101 |
| CEBPA | GCCCGGCAACTCTAGTATTA | TGACAAGGCACATATTTGCT | 102 |
| LIPE | CTTCTTCGAGGGTGATGAG | CGGGTGTGAACTGGAAAC | 107 |
| ACSF2 | GGCCATCAGCAGAGAAAGA | CACGCATGGTTGAGATGTC | 107 |
| BCKDHA | ACTTCGTCACCATCTCCT | ACAGATGACCACCCTGTT | 101 |
| GSTM3 | CTCACCTTTGTGGATTTCCTC | ACATGAAAGCCTTCAGATTCG | 100 |
| TMEM165 | AGATGAGTCCAGATGAAGGT | TCAACATCTCCTGGTCCATT | 112 |
| β-actin | GGATGCAGAAAGAGATCACT | TCTGCTGGAAGGTGGACA | 187 |
| Fatty Acid | Energy Stress (n = 6) | Control (n = 6) |
|---|---|---|
| C4:0 | 0.17 ± 0.04 a | 0.32 ± 0.15 b |
| C6:0 | 0.03 ± 0.01 | 0.05 ± 0.02 |
| C8:0 | 0.05 ± 0.03 A | 0.01 ± 0.00 B |
| C10:0 | 0.04 ± 0.01 | 0.04 ± 0.01 |
| C11:0 | 0.01 ± 0.00 | 0.01 ± 0.00 |
| C12:0 | 0.03 ± 0.00 | 0.03 ± 0.00 |
| C13:0 | 0.06 ± 0.03 | 0.13 ± 0.08 |
| C14:0 | 0.62 ± 0.13 | 0.57 ± 0.11 |
| C14:1 | 0.05 ± 0.01 | 0.04 ± 0.01 |
| C15:0 | 0.82 ± 0.03 A | 0.33 ± 0.04 B |
| C15:1 | 0.41 ± 0.06 A | 0.12 ± 0.03 B |
| C16:0 | 10.9 ± 0.68 A | 14.5 ± 2.23 B |
| C16:1 | 1.84 ± 0.12 A | 1.43 ± 0.16 B |
| C17:0 | 1.97 ± 0.13 A | 0.64 ± 0.18 B |
| C17:1 | 1.03 ± 0.10 A | 0.23 ± 0.13 B |
| C18:0 | 22.0 ± 0.87 A | 34.8 ± 2.04 B |
| cis-C18:1 | 12.9 ± 0.95 | 12.7 ± 1.19 |
| cis-C18:2 | 20.0 ± 1.74 | 19.4 ± 2.43 |
| C20:0 | 0.15 ± 0.02 | 0.08 ± 0.03 |
| C18:3n6 | 1.00 ± 0.17 A | 0.22 ± 0.13 B |
| C20:1n9 | 0.14 ± 0.04 A | 0.39 ± 0.11 B |
| C18:3n3 | 2.16 ± 0.45 A | 0.84 ± 0.20 B |
| C21:0 | 0.43 ± 0.09 A | 0.60 ± 0.13 B |
| C20:2 | 0.56 ± 0.32 a | 0.10 ± 0.04 b |
| C22:0 | 0.17 ± 0.11 | 0.15 ± 0.08 |
| C20:3n3 | 0.02 ± 0.00 A | 0.01 ± 0.00 B |
| C20:4n6 | 11.1 ± 0.56 A | 4.55 ± 0.38 B |
| C23:0 | 0.78 ± 0.28 A | 0.19 ± 0.06 B |
| C24:0 | 4.45 ± 0.36 | 4.19 ± 0.43 |
| C20:5n3 | 3.24 ± 0.64 A | 0.84 ± 0.30 B |
| C24:1 | 2.60 ± 0.42 | 2.54 ± 0.73 |
| C22:6n3 | 0.16 ± 0.07 A | 0.02 ± 0.01 B |
| SFA | 42.7 ± 1.46 A | 56.6 ± 0.73 B |
| MUFA | 19.0 ± 1.21 | 17.5 ± 1.83 |
| PUFA | 38.3 ± 1.33 A | 26.0 ± 2.43 B |
| UFA | 57.3 ± 1.46 A | 43.4 ± 0.73 B |
| SFA/UFA | 0.75 ± 0.05 A | 1.30 ± 0.04 B |
| MUFA/PUFA | 0.50 ± 0.04 a | 0.68 ± 0.13 b |
| PUFA/SFA | 0.90 ± 0.05 A | 0.46 ± 0.05 B |
| Item | Energy Stress (n = 6) | Control (n = 6) |
|---|---|---|
| GLU (mmol/L) | 3.84 ± 0.12 A | 4.95 ± 0.10 B |
| GC (ng/mL) | 89.2 ± 1.50 A | 50.3 ± 1.30 B |
| INS (ng/mL) | 9.53 ± 0.52 A | 12.0 ± 0.30 B |
| NEFA (nmol/L) | 174 ± 3.20 a | 180 ± 3.54 b |
| TG (mmol/L) | 0.19 ± 0.02 A | 0.25 ± 0.02 B |
| HDL (mmol/L) | 1.67 ± 0.12 | 1.77 ± 0.06 |
| LDL (mmol/L) | 0.60 ± 0.03 a | 0.65 ± 0.03 b |
| CH (mmol/L) | 2.59 ± 0.10 | 2.70 ± 0.06 |
| OA (mg/100 mL) | 13.6 ± 0.93 A | 11.2 ± 0.36 B |
| ACAC (mg/100 mL) | 31.8 ± 3.69 | 34.3 ± 2.47 |
| BHBA (mg/100 mL) | 4.26 ± 0.05 A | 2.15 ± 0.11 B |
| HSL (ng/mL) | 12.2 ± 1.38 a | 9.75 ± 0.18 b |
| ATGL (ng/mL) | 66.9 ± 1.28 A | 52.6 ± 1.38 B |
| ALB(g/L) | 26.5 ± 1.02 A | 31.2 ± 1.77 B |
| ID | Term | p | Score | ListHits |
|---|---|---|---|---|
| bom03010 | Ribosome | 1.23 × 10−21 | 2.50 | 99 |
| bom00190 | Oxidative phosphorylation | 9.98 × 10−16 | 2.68 | 63 |
| bom04714 | Thermogenesis | 1.45 × 10−13 | 2.14 | 88 |
| bom04146 | Peroxisome | 2.35 × 10−6 | 2.17 | 32 |
| bom04151 | PI3K-Akt signaling pathway | 3.21 × 10−5 | 1.47 | 91 |
| bom01040 | Biosynthesis of unsaturated fatty acids | 3.97 × 10−4 | 2.38 | 13 |
| bom04512 | ECM-receptor interaction | 6.69 × 10−4 | 2.77 | 27 |
| bom04152 | AMPK signaling pathway | 1.52 × 10−3 | 2.18 | 25 |
| bom00061 | Fatty acid biosynthesis | 7.56 × 10−3 | 2.22 | 7 |
| bom00790 | Folate biosynthesis | 8.43 × 10−3 | 1.95 | 10 |
| bom00071 | Fatty acid degradation | 1.15 × 10−2 | 1.74 | 13 |
| bom04923 | Regulation of lipolysis in adipocytes | 1.29 × 10−2 | 1.64 | 16 |
| bom04920 | Adipocytokine signaling pathway | 1.46 × 10−2 | 1.57 | 18 |
| bom00062 | Fatty acid elongation | 1.47 × 10−2 | 1.87 | 9 |
| bom00780 | Biotin metabolism | 1.59 × 10−2 | 3.02 | 2 |
| bom03320 | PPAR signaling pathway | 2.44 × 10−2 | 1.46 | 21 |
| bom04150 | mTOR signaling pathway | 2.55 × 10−2 | 1.33 | 35 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xiong, L.; Pei, J.; Wu, X.; Kalwar, Q.; Liang, C.; Guo, X.; Chu, M.; Bao, P.; Yao, X.; Yan, P. The Study of the Response of Fat Metabolism to Long-Term Energy Stress Based on Serum, Fatty Acid and Transcriptome Profiles in Yaks. Animals 2020, 10, 1150. https://doi.org/10.3390/ani10071150
Xiong L, Pei J, Wu X, Kalwar Q, Liang C, Guo X, Chu M, Bao P, Yao X, Yan P. The Study of the Response of Fat Metabolism to Long-Term Energy Stress Based on Serum, Fatty Acid and Transcriptome Profiles in Yaks. Animals. 2020; 10(7):1150. https://doi.org/10.3390/ani10071150
Chicago/Turabian StyleXiong, Lin, Jie Pei, Xiaoyun Wu, Qudratullah Kalwar, Chunnian Liang, Xian Guo, Min Chu, Pengjia Bao, Xixi Yao, and Ping Yan. 2020. "The Study of the Response of Fat Metabolism to Long-Term Energy Stress Based on Serum, Fatty Acid and Transcriptome Profiles in Yaks" Animals 10, no. 7: 1150. https://doi.org/10.3390/ani10071150
APA StyleXiong, L., Pei, J., Wu, X., Kalwar, Q., Liang, C., Guo, X., Chu, M., Bao, P., Yao, X., & Yan, P. (2020). The Study of the Response of Fat Metabolism to Long-Term Energy Stress Based on Serum, Fatty Acid and Transcriptome Profiles in Yaks. Animals, 10(7), 1150. https://doi.org/10.3390/ani10071150





