Luminal and Mucosal Microbiota of the Cecum and Large Colon of Healthy and Diarrheic Horses
Abstract
:Simple Summary
Abstract
1. Introduction
2. Material and Methods
2.1. Animals
Healthy Horses
2.2. Colitis Cases
2.3. Samples Collection
2.4. DNA Extraction and Sequencing of the V3-V4 Region of the 16S rRNA Gene
2.5. Sequence Processing and Data Analysis
3. Results
3.1. Analysis of 16S rRNA Gene Sequencing
3.2. Alpha Diversity
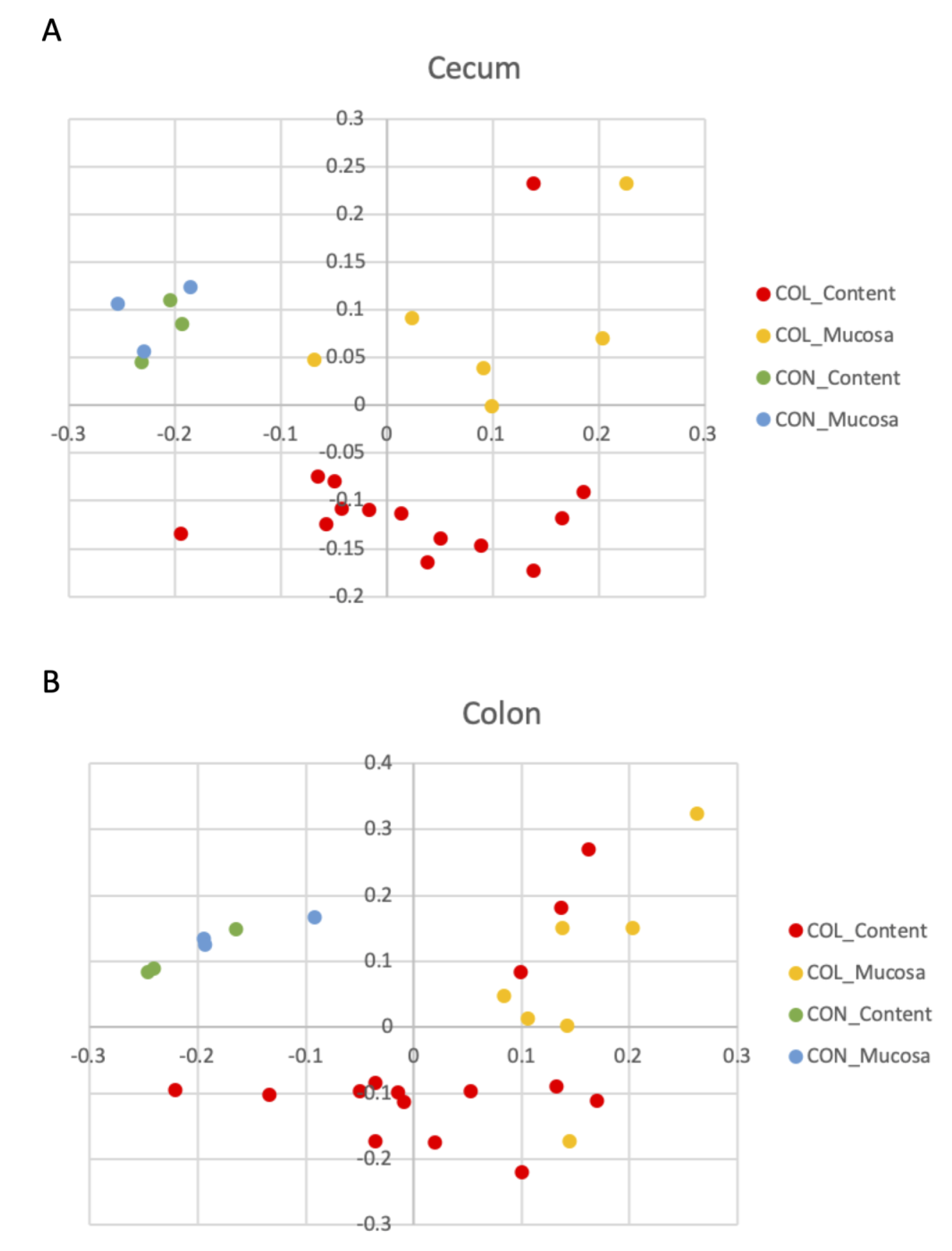
3.3. Beta Diversity
3.4. Relative Abundance and LefSe Analysis
4. Discussion
4.1. Comparison between Mucosal and Luminal Content
4.2. Comparison between Cecum and Large Colon
4.3. Comparison between Healthy and Colitis Cases
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Shaw, S.D.; Stämpfli, H.R. Diagnosis and Treatment of Undifferentiated and Infectious Acute Diarrhea in the Adult Horse. Vet. Clin. Equine Pract. 2018, 34, 39–53. [Google Scholar] [CrossRef] [PubMed]
- Uzal, F.A.; Diab, S.S. Gastritis, Enteritis, and Colitis in Horses. Vet. Clin. Equine Pract. 2015, 31, 337–358. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feary, D.J.; Hassel, D.M. Enteritis and colitis in horses. Vet. Clin. Equine Pract. 2006, 22, 437–479. [Google Scholar] [CrossRef] [PubMed]
- Macías-Rioseco, M.; Hill, A.E.; Uzal, F.A. Fatal intestinal inflammatory lesions in equids in California: 710 cases (1990–2013). J. Am. Vet. Med. Assoc. 2020, 256, 455–462. [Google Scholar] [CrossRef] [PubMed]
- Costa, M.C.; Weese, J.S. The equine intestinal microbiome. Anim. Health Res. Rev. 2012, 13, 121–128. [Google Scholar] [CrossRef]
- Costa, M.C.; Weese, J.S. Understanding the Intestinal Microbiome in Health and Disease. Vet. Clin. N. Am. Equine Pract. 2018, 34, 1–12. [Google Scholar] [CrossRef]
- Costa, M.C.; Silva, G.; Ramos, R.V.; Staempfli, H.R.; Arroyo, L.G.; Kim, P.; Weese, J.S. Characterization and comparison of the bacterial microbiota in different gastrointestinal tract compartments in horses. Vet. J. 2015, 205, 74–80. [Google Scholar] [CrossRef]
- Ni, J.; Wu, G.; Albenberg, L.; Vesselin, T.T. Gut microbiota and IBD: Causation or correlation? Nat. Rev. Gastroenterol. Hepatol. 2017, 14, 573–584. [Google Scholar] [CrossRef] [Green Version]
- Kiely, C.J.; Pavli, P.; O’Brien, C.L. The microbiome of translocated bacterial populations in patients with and without inflammatory bowel disease. Intern. Med. J. 2018, 48, 1346–1354. [Google Scholar] [CrossRef]
- Rodriguez, C.; Taminiau, B.; Brévers, B.; Avesani, V.; Van Broeck, J.; Leroux, A.; Gallot, M.; Bruwier, A.; Amory, H.; Delmée, M.; et al. Faecal microbiota characterisation of horses using 16 rDNA barcoded pyrosequencing, and carriage rate of Clostridium difficile at hospital admission. BMC Microbiol. 2015, 15, 181. [Google Scholar] [CrossRef] [Green Version]
- Schoster, A.; Staempfli, H.R.; Guardabassi, L.G.; Jalali, M.; Weese, J.S. Comparison of the fecal bacterial microbiota of healthy and diarrheic foals at two and four weeks of life. BMC Vet. Res. 2017, 13, 144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Costa, M.C.; Arroyo, L.G.; Allen-Vercoe, E.; Stämpfli, H.R.; Kim, P.T.; Sturgeon, A.; Weese, J.S. Comparison of the fecal microbiota of healthy horses and horses with colitis by high throughput sequencing of the V3–V5 region of the 16S rRNA gene. PLoS ONE 2012, 7, e41484. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ericsson, A.C.; Johnson, P.J.; Lopes, M.A.; Perry, S.C.; Lanter, H.R. A Microbiological Map of the Healthy Equine Gastrointestinal Tract. PLoS ONE 2016, 11, e0166523. [Google Scholar] [CrossRef] [PubMed]
- Ringel, Y.; Maharshak, N.; Ringel-Kulka, T.; Wolber, E.A.; Sartor, R.B.; Carroll, I.M. High throughput sequencing reveals distinct microbial populations within the mucosal and luminal niches in healthy individuals. Gut Microbes 2015, 6, 173–181. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mottawea, W.; Butcher, J.; Li, J.; Abujamel, T.; Manoogian, J.; Mack, D.; Stintzi, A. The mucosal-luminal interface: An ideal sample to study the mucosa-associated microbiota and the intestinal microbial biogeography. Pediatr. Res. 2019, 85, 895–903. [Google Scholar] [CrossRef]
- De Weirdt, R.; Van de Wiele, T. Micromanagement in the gut: Microenvironmental factors govern colon mucosal biofilm structure and functionality. NPJ Biofilm. Microbiomes 2015, 1, 15026. [Google Scholar] [CrossRef] [Green Version]
- Integrative HMP (iHMP) Research Network Consortium. The Integrative Human Microbiome Project: Dynamic analysis of microbiome-host omics profiles during periods of human health and disease. Cell Host Microbe 2014, 10, 276–289. [Google Scholar]
- Lynch, S.V.; Pedersen, O. The Human Intestinal Microbiome in Health and Disease. N. Engl. J. Med. 2016, 15, 2369–2379. [Google Scholar] [CrossRef] [Green Version]
- Thomas, S.; Izard, J.; Walsh, E.; Batich, K.; Chongsathidkiet, P.; Clarke, G.; Sela, D.A.; Muller, A.J.; Mullin, J.M.; Albert, K.; et al. The Host Microbiome Regulates and Maintains Human Health: A Primer and Perspective for Non-Microbiologists. Cancer Res. 2017, 15, 1783–1812. [Google Scholar] [CrossRef] [Green Version]
- Youmans, B.P.; Ajami, N.J.; Jiang, Z.D.; Campbell, F.; Wadsworth, W.D.; Petrosino, J.F.; DuPont, H.L.; Highlander, S.K. Characterization of the human gut microbiome during travelers’ diarrhea. Gut Microbes 2015, 6, 110–119. [Google Scholar] [CrossRef]
- David, L.A.; Weil, A.; Ryan, E.T.; Calderwood, S.B.; Harris, J.B.; Chowdhury, F.; Begum, Y.; Qadri, F.; LaRocque, R.C.; Turnbaugh, P.J. Gut microbial succession follows acute secretory diarrhea in humans. mBio 2015, 19, e00381-15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klindworth, A.; Pruesse, E.; Schweer, T.; Peplies, J.; Quast, C.; Horn, M.; Glöckner, F.O. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013, 7, e1. [Google Scholar] [CrossRef] [PubMed]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kozich, J.J.; Westcott, S.L.; Baxter, N.T.; Highlander, S.K.; Schloss, P.D. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl. Environ. Microbiol. 2013, 79, 5112–5120. [Google Scholar] [CrossRef] [Green Version]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2012, 41, D590–D596. [Google Scholar] [CrossRef]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, 1–18. [Google Scholar] [CrossRef] [Green Version]
- Dowds, C.M.; Blumberg, R.S.; Zeissig, S. Control of intestinal homeostasis through crosstalk between natural killer T cells and the intestinal microbiota. Clin. Immunol. 2015, 159, 128–133. [Google Scholar] [CrossRef] [Green Version]
- Gensollen, T.; Iyer, S.S.; Kasper, D.L.; Blumberg, R.S. How colonization by microbiota in early life shapes the immune system. Science 2016, 29, 539–544. [Google Scholar] [CrossRef] [Green Version]
- Yarandi, S.S.; Kulkarni, S.; Saha, M.; Sylvia, K.E.; Sears, C.L.; Pasricha, P.J. Intestinal Bacteria Maintain Adult Enteric Nervous System and Nitrergic Neurons via Toll-like Receptor 2-induced Neurogenesis in Mice. Gastroenterology 2020, 159, 200–213.e8. [Google Scholar] [CrossRef]
- Araújo-Pérez, F.; McCoy, A.N.; Okechukwu, C.; Carroll, I.M.; Smith, K.M.; Jeremiah, K.; Sandler, R.S.; Asher, G.N.; Keku, T.O. Differences in microbial signatures between rectal mucosal biopsies and rectal swabs. Gut Microbes 2012, 3, 530–535. [Google Scholar] [CrossRef] [Green Version]
- Oliphant, K.; Parreira, V.R.; Cochrane, K.; Allen-Vercoe, E. Drivers of human gut microbial community assembly: Coadaptation, determinism and stochasticity. ISME J. 2019, 13, 3080–3092. [Google Scholar] [CrossRef] [PubMed]
- Dougal, K.; Harris, P.A.; Edwards, A.; Pachebat, J.A.; Blackmore, T.M.; Worgan, H.J.; Newbold, C.J. A comparison of the microbiome and the metabolome of different regions of the equine hindgut. FEMS Microbiol. Ecol. 2012, 82, 642–652. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salem, S.E.; Maddox, T.W.; Antczak, P.; Ketley, J.M.; Williams, N.J.; Archer, D.C. Acute changes in the colonic microbiota are associated with large intestinal forms of surgical colic. BMC Vet. Res. 2019, 15, 468. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Devkota, S.; Wang, Y.; Musch, M.W.; Leone, V.; Fehlner-Peach, H.; Nadimpalli, A.; Antonopoulos, D.A.; Jabri, B.; Chang, E.B. Dietary-fat-induced taurocholic acid promotes pathobiont expansion and colitis in Il10-/- mice. Nature 2012, 5, 104–108. [Google Scholar] [CrossRef] [Green Version]
- Maharshak, N.; Packey, C.D.; Ellermann, M.; Manick, S.; Siddle, J.P.; Huh, E.Y.; Plevy, S.; Sartor, R.B.; Carroll, I.M. Altered enteric microbiota ecology in interleukin 10-deficient mice during development and progression of intestinal inflammation. Gut Microbes 2013, 4, 316–324. [Google Scholar] [CrossRef]
- Winter, S.E.; Winter, M.G.; Xavier, M.N.; Thiennimitr, P.; Poon, V.; Keestra, A.M.; Laughlin, R.C.; Gomez, G.; Wu, J.; Lawhon, S.D.; et al. Host-derived nitrate boosts growth of E. coli in the inflamed gut. Science 2013, 8, 708–711. [Google Scholar] [CrossRef] [Green Version]
- Gomez, D.E.; Arroyo, L.G.; Costa, M.C.; Viel, L.; Weese, J.S. Characterization of the Fecal Bacterial Microbiota of Healthy and Diarrheic Dairy Calves. J. Vet. Intern. Med. 2017, 31, 928–939. [Google Scholar] [CrossRef]
- Stecher, B.; Chaffron, S.; Käppeli, R.; Hapfelmeier, S.; Freedrich, S.; Weber, T.C.; Kirundi, J.; Suar, M.; McCoy, K.D.; von Mering, C.; et al. Like will to like: Abundances of closely related species can predict susceptibility to intestinal colonization by pathogenic and commensal bacteria. PLoS Pathog. 2010, 6, e1000711. [Google Scholar] [CrossRef]
- Shin, N.R.; Whon, T.W.; Bae, J.W. Proteobacteria: Microbial signature of dysbiosis in gut microbiota. Trends Biotechnol. 2015, 33, 496–503. [Google Scholar] [CrossRef]
- McKinney, C.A.; Oliveira, B.C.M.; Bedenice, D.; Paradis, M.R.; Mazan, M.; Sage, S.; Sanchez, A.; Widmer, G. The fecal microbiota of healthy donor horses and geriatric recipients undergoing fecal microbial transplantation for the treatment of diarrhea. PLoS ONE 2020, 15, e0230148. [Google Scholar] [CrossRef]



| Bred | Age (Years) | Sex | Body Weight (Kg) | Histophatological Diagnosis | Antibiotics Administered |
|---|---|---|---|---|---|
| QH | 4 | MC | 345 | Fibrinonecrotic typhlocolitis | TMS, Pen, Gen |
| TB | 4 | Male | 538 | Segmental ulcerative colitis | Yes, unknown |
| TB | 19 | MC | 528 | Fibrinonecrotic typhlocolitis | TMS |
| STB | 4 | MC | 536 | Colonic edema | Pen, Gen |
| Belgian | 21 | F | 702 | Transmural necrosis cecum and colon | Pen, Gen, Metro |
| TB | 1 | F | 380 | L. intracellularis enteritis and necrotizing colitis | Oxytetetracycline |
| MB | 6 | F | N/A | Necrotizing and hemorrhagic ulcerative colitis | TMS, Metro, Gen, pen |
| p-Value | df and (MS) | p-Value | df and (MS) | |
|---|---|---|---|---|
| Cecum | Colon | |||
| Colitis—Healthy (content) | 0.003 | 16 (0.272, 0.151) | 0.008 | 17 (0.248, 0.149) |
| Colitis—Healthy (mucosa) | 0.010 | 8 (0.301, 0.109) | 0.002 | 9 (0.290, 0.136) |
| Content—Mucosa (Colitis) | 0.003 | 19 (0.246, 0.151) | 0.034 | 21 (0.242, 0.155) |
| Content—Mucosa (Healthy) | 0.108 | 5 (0.137, 0.079) | 0.143 | 5 (0.124, 0.094) |
| Healthy | Colitis | |||
| Cecum—Colon (Content) | 0.673 | 5 (0.070, 0.084) | 0.981 | 28 (0.097, 0.160) |
| Cecum—Colon (Mucosa) | 0.615 | 5 (0.089, 0.089) | 0.988 | 12 (0.084, 0.136) |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Arroyo, L.G.; Rossi, L.; Santos, B.P.; Gomez, D.E.; Surette, M.G.; Costa, M.C. Luminal and Mucosal Microbiota of the Cecum and Large Colon of Healthy and Diarrheic Horses. Animals 2020, 10, 1403. https://doi.org/10.3390/ani10081403
Arroyo LG, Rossi L, Santos BP, Gomez DE, Surette MG, Costa MC. Luminal and Mucosal Microbiota of the Cecum and Large Colon of Healthy and Diarrheic Horses. Animals. 2020; 10(8):1403. https://doi.org/10.3390/ani10081403
Chicago/Turabian StyleArroyo, Luis G., Laura Rossi, Bruna P Santos, Diego E Gomez, Michael G Surette, and Marcio C Costa. 2020. "Luminal and Mucosal Microbiota of the Cecum and Large Colon of Healthy and Diarrheic Horses" Animals 10, no. 8: 1403. https://doi.org/10.3390/ani10081403





