Genomic Evaluation of Primiparous High-Producing Dairy Cows: Inbreeding Effects on Genotypic and Phenotypic Production–Reproductive Traits
Abstract
:Simple Summary
Abstract
1. Introduction
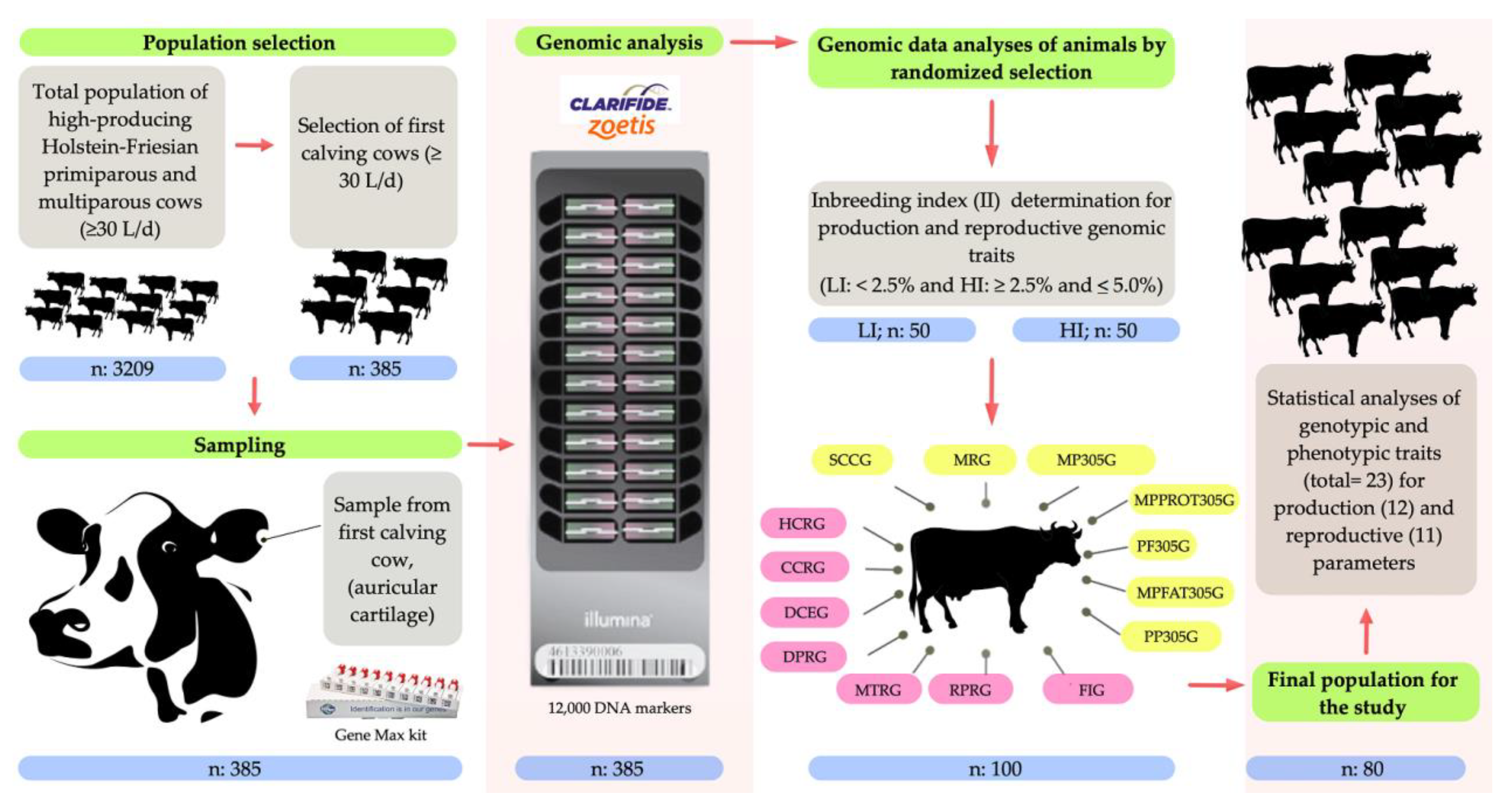
2. Materials and Methods
2.1. Ethical Statement
2.2. Location, Environmental Conditions, and Animals
2.3. Genomic Analysis
2.4. Analysis of the Inbreeding Index (II)
2.5. Analysis of Genomic Parameters for Production Traits
2.6. Analysis of Genomic Parameters for Reproductive Traits
2.7. Analysis of Phenotypic Parameters for Production Traits
2.8. Analysis of Phenotypic Parameters for Reproductive Traits
2.9. Statistical Analysis
3. Results
3.1. Analysis of Genomic Parameters for Production Traits
3.2. Analysis of Phenotypic Parameters for Production Traits
3.3. Analysis of Genomic Parameters for Reproductive Traits
3.4. Analysis of Phenotypic Parameters for Reproductive Traits
3.5. Linear Correlation Analyses between Genomic Parameters of Production and Reproductive Traits
3.6. Linear Correlation Analyses within Genomic Parameters of Production and Reproductive Traits
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- García-Ruiz, A.; Cole, J.B.; VanRaden, P.M.; Wiggans, G.R.; Ruiz-López, F.J.; Van Tassell, C.P. Changes in genetic selection differentials and generation intervals in US Holstein dairy cattle as a result of genomic selection. Proc. Natl. Acad. Sci. USA 2016, 113, E3995–E4004. [Google Scholar] [CrossRef] [Green Version]
- Weller, J.I.; Ezra, E.; Ron, M. Invited review: A perspective on the future of genomic selection in dairy cattle. J. Dairy Sci. 2017, 100, 8633–8644. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wiggans, G.R.; Vanraden, P.M.; Cooper, T.A. The genomic evaluation system in the United States: Past, present, future. J. Dairy Sci. 2011, 94, 3202–3211. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thomasen, J.R.; Egger-Danner, C.; Willam, A.; Guldbrandtsen, B.; Lund, M.S.; Sørensen, A.C. Genomic selection strategies in a small dairy cattle population evaluated for genetic gain and profit. J. Dairy Sci. 2014, 97, 458–470. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pryce, J.E.; Nguyen, T.T.T.; Axford, M.; Nieuwhof, G.; Shaffer, M. Symposium review: Building a better cow—The Australian experience and future perspectives. J. Dairy Sci. 2018, 101, 3702–3713. [Google Scholar] [CrossRef] [PubMed]
- Misztal, I.; Legarra, A. Invited review: Efficient computation strategies in genomic selection. Animal 2017, 11, 731–736. [Google Scholar] [CrossRef] [Green Version]
- Jenko, J.; Wiggans, G.R.; Cooper, T.A.; Eaglen, S.A.E.; Luff, W.G.d.L.; Bichard, M.; Pong-Wong, R.; Woolliams, J.A. Cow genotyping strategies for genomic selection in a small dairy cattle population. J. Dairy Sci. 2017, 100, 439–452. [Google Scholar] [CrossRef] [Green Version]
- Wiggans, G.R.; Cole, J.B.; Hubbard, S.M.; Sonstegard, T.S. Genomic Selection in Dairy Cattle: The USDA Experience. Annu. Rev. Anim. Biosci. 2017, 5, 309–327. [Google Scholar] [CrossRef] [Green Version]
- Howard, J.T.; Pryce, J.E.; Baes, C.; Maltecca, C. Invited review: Inbreeding in the genomics era: Inbreeding, inbreeding depression, and management of genomic variability. J. Dairy Sci. 2017, 100, 6009–6024. [Google Scholar] [CrossRef]
- Hermas, S.A.; Young, C.W.; Rust, J.W. Effects of Mild Inbreeding on Productive and Reproductive Performance of Guernsey Cattle. J. Dairy Sci. 1987, 70, 712–715. [Google Scholar] [CrossRef]
- Egger-Danner, C.; Cole, J.B.; Pryce, J.E.; Gengler, N.; Heringstad, B.; Bradley, A.; Stock, K.F. Invited review: Overview of new traits and phenotyping strategies in dairy cattle with a focus on functional traits. Animal 2015, 9, 191–207. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reiner-Benaim, A.; Ezra, E.; Weller, J.I. Optimization of a genomic breeding program for a moderately sized dairy cattle population. J. Dairy Sci. 2017, 100, 2892–2904. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schöpke, K.; Swalve, H.H. Review: Opportunities and challenges for small populations of dairy cattle in the era of genomics. Animal 2016, 10, 1050–1060. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Haas, Y.; Pszczola, M.; Soyeurt, H.; Wall, E.; Lassen, J. Invited review: Phenotypes to genetically reduce greenhouse gas emissions in dairying. J. Dairy Sci. 2017, 100, 855–870. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mucha, A.; Wierzbicki, H.; Kamiński, S.; Oleński, K.; Hering, D. High-frequency marker haplotypes in the genomic selection of dairy cattle. J. Appl. Genet. 2019, 60, 179–186. [Google Scholar] [CrossRef] [Green Version]
- Mc Parland, S.; Kearney, J.F.; Rath, M.; Berry, D.P. Inbreeding effects on milk production, calving performance, fertility, and conformation in Irish Holstein-Friesians. J. Dairy Sci. 2007, 90, 4411–4419. [Google Scholar] [CrossRef]
- Koivula, M.; Strandén, I.; Aamand, G.P.; Mäntysaari, E.A. Effect of cow reference group on validation reliability of genomic evaluation. Animal 2016, 10, 1061–1066. [Google Scholar] [CrossRef] [Green Version]
- Mäntysaari, E.A.; Strandén, I. Genomic data and breeding value estimation in dairy cattle: Theory, practice, problems. J. Anim. Breed. Genet. 2016, 133, 165–166. [Google Scholar] [CrossRef]
- Weigel, K.A.; Lin, S.W. Use of computerized mate selection programs to control inbreeding of Holstein and Jersey cattle in the next generation. J. Dairy Sci. 2000, 83, 822–828. [Google Scholar] [CrossRef]
- Macedo, A.A.; Bittar, J.F.F.; Bassi, P.B.; Ronda, J.B.; Bittar, E.R.; Panetto, J.C.C.; Araujo, M.S.S.; Santos, R.L.; Martins-Filho, O.A. Influence of endogamy and mitochondrial DNA on immunological parameters in cattle. BMC Vet. Res. 2014, 10, 79. [Google Scholar] [CrossRef] [Green Version]
- Moore, S.G.; Hasler, J.F. A 100-Year Review: Reproductive technologies in dairy science. J. Dairy Sci. 2017, 100, 10314–10331. [Google Scholar] [CrossRef] [PubMed]
- Aguilar, I.; Misztal, I.; Johnson, D.L.; Legarra, A.; Tsuruta, S.; Lawlor, T.J. Hot topic: A unified approach to utilize phenotypic, full pedigree, and genomic information for genetic evaluation of Holstein final score. J. Dairy Sci. 2010, 93, 743–752. [Google Scholar] [CrossRef]
- Taylor, J.F.; Schnabel, R.D.; Sutovsky, P. Review: Genomics of bull fertility. Animal 2018, 12, s172–s183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, L.; Bickhart, D.M.; Cole, J.B.; Schroeder, S.G.; Song, J.; Van Tassell, C.P.; Sonstegard, T.S.; Liu, G.E. Genomic signatures reveal new evidences for selection of important traits in domestic cattle. Mol. Biol. Evol. 2015, 32, 711–725. [Google Scholar] [CrossRef] [Green Version]
- Maiwashe, A.; Nephawe, K.A.; Theron, H.E. Estimates of genetic parameters and effect of inbreeding on milk yield and composition in South African Jersey cows. S. Afr. J. Anim. Sci. 2008, 38, 119–125. [Google Scholar] [CrossRef]
- Mc Parland, S.; Kearney, F.; Berry, D.P. Purging of inbreeding depression within the Irish Holstein-Friesian population. Genet. Sel. Evol. 2009, 41, 16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rokouei, M.; Vaez Torshizi, R.; Moradi Shahrbabak, M.; Sargolzaei, M.; Sørensen, A.C. Monitoring inbreeding trends and inbreeding depression for economically important traits of Holstein cattle in Iran. J. Dairy Sci. 2010, 93, 3294–3302. [Google Scholar] [CrossRef] [PubMed]
- Pryce, J.E.; Haile-Mariam, M.; Goddard, M.E.; Hayes, B.J. Identification of genomic regions associated with inbreeding depression in Holstein and Jersey dairy cattle. Genet. Sel. Evol. 2014, 46, 71. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dezetter, C.; Leclerc, H.; Mattalia, S.; Barbat, A.; Boichard, D.; Ducrocq, V. Inbreeding and crossbreeding parameters for production and fertility traits in Holstein, Montbéliarde, and Normande cows. J. Dairy Sci. 2015, 98, 4904–4913. [Google Scholar] [CrossRef] [Green Version]
- Doekes, H.P.; Veerkamp, R.F.; Bijma, P.; De Jong, G.; Hiemstra, S.J.; Windig, J.J. Inbreeding depression due to recent and ancient inbreeding in Dutch Holstein-Friesian dairy cattle. Genet. Sel. Evol. 2019, 51, 54. [Google Scholar] [CrossRef] [Green Version]
- Martikainen, K.; Koivula, M.; Uimari, P. Identification of runs of homozygosity affecting female fertility and milk production traits in Finnish Ayrshire cattle. Sci. Rep. 2020, 10, 3804. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.R.; Everett, R.W.; Wolfe, C.W. Effects of inbreeding on production and survival in Jerseys. J. Dairy Sci. 2000, 83, 2131–2138. [Google Scholar] [CrossRef]
- Smith, L.A.; Cassell, B.G.; Pearson, R.E. The Effects of Inbreeding on the Lifetime Performance of Dairy Cattle. J. Dairy Sci. 1998, 81, 2729–2737. [Google Scholar] [CrossRef]
- Berry, D.P.; Wall, E.; Pryce, J.E. Genetics and genomics of reproductive performance in dairy and beef cattle. Animal 2014, 8, 105–121. [Google Scholar] [CrossRef] [Green Version]
- Miller, N.; Delbecchi, L.; Petitclerc, D.; Wagner, G.F.; Talbot, B.G.; Lacasse, P. Effect of stage of lactation and parity on mammary gland cell renewal. J. Dairy Sci. 2006, 89, 4669–4677. [Google Scholar] [CrossRef]
- Kessler, E.C.; Bruckmaier, R.M.; Gross, J.J. Milk production during the colostral period is not related to the later lactational performance in dairy cows. J. Dairy Sci. 2014, 97, 2186–2192. [Google Scholar] [CrossRef] [Green Version]
- Bjelland, D.W.; Weigel, K.A.; Vukasinovic, N.; Nkrumah, J.D. Evaluation of inbreeding depression in Holstein cattle using whole-genome SNP markers and alternative measures of genomic inbreeding. J. Dairy Sci. 2013, 96, 4697–4706. [Google Scholar] [CrossRef]
- Battagin, M.; Sartori, C.; Biffani, S.; Penasa, M.; Cassandro, M. Genetic parameters for body condition score, locomotion, angularity, and production traits in Italian Holstein cattle. J. Dairy Sci. 2013, 96, 5344–5351. [Google Scholar] [CrossRef] [Green Version]
- Baes, C.F.; Makanjuola, B.O.; Miglior, F.; Marras, G.; Howard, J.T.; Fleming, A.; Maltecca, C. Symposium review: The genomic architecture of inbreeding: How homozygosity affects health and performance. J. Dairy Sci. 2019, 102, 2807–2817. [Google Scholar] [CrossRef]
- Miglior, F.; Van Doormaal, B.J.; Kistemaker, G.; Canada, A.; Network, C.D.; Swiss, B. Phenotypic analysis of inbreeding depression for traits measured in Canadian dairy cattle breeds. Can. Dairy Netw. 2001, 1967, 1–16. [Google Scholar]
- Makanjuola, B.O.; Maltecca, C.; Miglior, F.; Schenkel, F.S.; Baes, C.F. Effect of recent and ancient inbreeding on production and fertility traits in Canadian Holsteins. BMC Genom. 2020, 21, 605. [Google Scholar] [CrossRef] [PubMed]
- Leroy, G. Inbreeding depression in livestock species: Review and meta-analysis. Anim. Genet. 2014, 45, 618–628. [Google Scholar] [CrossRef] [PubMed]
- Oikonomou, G.; Arsenos, G.; Valergakis, G.E.; Tsiaras, A.; Zygoyiannis, D.; Banos, G. Genetic relationship of body energy and blood metabolites with reproduction in Holstein cows. J. Dairy Sci. 2008, 91, 4323–4332. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bastin, C.; Laloux, L.; Gillon, A.; Miglior, F.; Soyeurt, H.; Hammami, H.; Bertozzi, C.; Gengler, N. Modeling milk urea of Walloon dairy cows in management perspectives. J. Dairy Sci. 2009, 92, 3529–3540. [Google Scholar] [CrossRef] [Green Version]
- Hossein-Zadeh, N.G.; Ardalan, M. Genetic relationship between milk urea nitrogen and reproductive performance in Holstein dairy cows. Animal 2011, 5, 26–32. [Google Scholar] [CrossRef] [Green Version]
- Rzewuska, K.; Strabel, T. Genetic parameters for milk urea concentration and milk traits in Polish Holstein-Friesian cows. J. Appl. Genet. 2013, 54, 473–482. [Google Scholar] [CrossRef] [Green Version]
- Hinrichs, D.; Thaller, G. Pedigree analysis and inbreeding effects on calving traits in large dairy herds in Germany. J. Dairy Sci. 2011, 94, 4726–4733. [Google Scholar] [CrossRef]
- Fritz, S.; Capitan, A.; Djari, A.; Rodriguez, S.C.; Barbat, A.; Baur, A.; Grohs, C.; Weiss, B.; Boussaha, M.; Esquerré, D.; et al. Detection of Haplotypes Associated with Prenatal Death in Dairy Cattle and Identification of Deleterious Mutations in GART, SHBG and SLC37A2. PLoS ONE 2013, 8, e65550. [Google Scholar] [CrossRef]
- Wu, X.; Mesbah-Uddin, M.; Guldbrandtsen, B.; Lund, M.S.; Sahana, G. Novel haplotypes responsible for prenatal death in Nordic Red and Danish Jersey cattle. J. Dairy Sci. 2020, 103, 4570–4578. [Google Scholar] [CrossRef] [Green Version]
- Kindahl, H.; Kornmatitsuk, B.; Königsson, K.; Gustafsson, H. Endocrine changes in late bovine pregnancy with special emphasis on fetal well-being. Domest. Anim. Endocrinol. 2002, 23, 321–328. [Google Scholar] [CrossRef]
- VanRaden, P.M.; Miller, R.H. Effects of nonadditive genetic interactions, inbreeding, and recessive defects on embryo and fetal loss by seventy days. J. Dairy Sci. 2006, 89, 2716–2721. [Google Scholar] [CrossRef]
- Martikainen, K.; Tyrisevä, A.M.; Matilainen, K.; Pösö, J.; Uimari, P. Estimation of inbreeding depression on female fertility in the Finnish Ayrshire population. J. Anim. Breed. Genet. 2017, 134, 383–392. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perez, B.C.; Balieiro, J.C.C.; Ventura, R.V.; Bruneli, F.A.T.; Peixoto, M.G.C.D. Inbreeding effects on in vitro embryo production traits in Guzerá cattle. Animal 2017, 11, 1983–1990. [Google Scholar] [CrossRef]
- Heersche, G.; Nebel, R.L. Measuring Efficiency and Accuracy of Detection of Estrus. J. Dairy Sci. 1994, 77, 2754–2761. [Google Scholar] [CrossRef]
- Palmer, M.A.; Olmos, G.; Boyle, L.A.; Mee, J.F. Estrus detection and estrus characteristics in housed and pastured Holstein-Friesian cows. Theriogenology 2010, 74, 255–264. [Google Scholar] [CrossRef]
- Saint-Dizier, M.; Chastant-Maillard, S. Towards an Automated Detection of Oestrus in Dairy Cattle. Reprod. Domest. Anim. 2012, 47, 1056–1061. [Google Scholar] [CrossRef] [Green Version]
- Dobson, H.; Williams, J.; Routly, J.E.; Jones, D.N.; Cameron, J.; Holman-Coates, A.; Smith, R.F. Short communication: Chronology of different sexual behaviors and motion activity during estrus in dairy cows. J. Dairy Sci. 2018, 101, 8291–8295. [Google Scholar] [CrossRef]
- Marques, O.; Veronese, A.; Merenda, V.R.; Bisinotto, R.S.; Chebel, R.C. Effect of estrous detection strategy on pregnancy outcomes of lactating Holstein cows receiving artificial insemination and embryo transfer. J. Dairy Sci. 2020, 103, 6635–6646. [Google Scholar] [CrossRef]
- Pfeiffer, J.; Gandorfer, M.; Ettema, J.F. Evaluation of activity meters for estrus detection: A stochastic bioeconomic modeling approach. J. Dairy Sci. 2020, 103, 492–506. [Google Scholar] [CrossRef]
- Reith, S.; Hoy, S. Review: Behavioral signs of estrus and the potential of fully automated systems for detection of estrus in dairy cattle. Animal 2018, 12, 398–407. [Google Scholar] [CrossRef] [Green Version]
- Doublet, A.C.; Croiseau, P.; Fritz, S.; Michenet, A.; Hozé, C.; Danchin-Burge, C.; Laloë, D.; Restoux, G. The impact of genomic selection on genetic diversity and genetic gain in three French dairy cattle breeds. Genet. Sel. Evol. 2019, 51, 52. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huson, H.J.; Sonstegard, T.S.; Godfrey, J.; Hambrook, D.; Wolfe, C.; Wiggans, G.; Blackburn, H.; VanTassell, C.P. A Genetic Investigation of Island Jersey Cattle, the Foundation of the Jersey Breed: Comparing Population Structure and Selection to Guernsey, Holstein, and United States Jersey Cattle. Front. Genet. 2020, 11, 366. [Google Scholar] [CrossRef] [PubMed]
- Mao, X.; Sahana, G.; Johansson, A.M.; Liu, A.; Ismael, A.; Løvendahl, P.; De Koning, D.J.; Guldbrandtsen, B. Genome-wide association mapping for dominance effects in female fertility using real and simulated data from Danish Holstein cattle. Sci. Rep. 2020, 10, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Ilska-Warner, J.J.; Psifidi, A.; Seeker, L.A.; Wilbourn, R.V.; Underwood, S.L.; Fairlie, J.; Whitelaw, B.; Nussey, D.H.; Coffey, M.P.; Banos, G. The Genetic Architecture of Bovine Telomere Length in Early Life and Association With Animal Fitness. Front. Genet. 2019, 10, 1048. [Google Scholar] [CrossRef] [Green Version]
- Buttchereit, N.; Stamer, E.; Junge, W.; Thaller, G. Genetic parameters for energy balance, fat/protein ratio, body condition score and disease traits in German Holstein cows. J. Anim. Breed. Genet. 2012, 129, 280–288. [Google Scholar] [CrossRef]
- Benedet, A.; Manuelian, C.L.; Penasa, M.; Cassandro, M.; Righi, F.; Sternieri, M.; Galimberti, P.; Zambrini, A.V.; De Marchi, M. Factors associated with herd bulk milk composition and technological traits in the Italian dairy industry. J. Dairy Sci. 2018, 101, 934–943. [Google Scholar] [CrossRef] [Green Version]




| Genotypic Parameters | FIG (pts) | DPRG (%) | HCRG (%) | CCRG (%) | DCEG (%) | MTRG (%) | RPRG (%) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| r | p | r | p | r | p | r | p | r | p | r | p | r | p | |
| MP305G (lbs) | −0.476 | 0.034 | −0.565 | 0.010 | −0.217 | NS | −0.250 | NS | 0.027 | NS | −0.128 | NS | 0.025 | NS |
| MPFAT305G (lbs) | −0.223 | NS | −0.237 | NS | 0.017 | NS | −0.405 | NS | −0.222 | NS | 0.338 | NS | −0.123 | NS |
| MPPROT305G (lbs) | −0.517 | 0.020 | −0.593 | 0.006 | −0.242 | NS | −0.355 | NS | 0.168 | NS | −0.002 | NS | 0.065 | NS |
| PF305G (%) | 0.260 | NS | 0.328 | NS | 0.199 | NS | −0.063 | NS | −0.170 | NS | 0.356 | NS | −0.089 | NS |
| PP305G (%) | 0.078 | NS | 0.130 | NS | −0.001 | NS | −0.078 | NS | 0.210 | NS | 0.202 | NS | 0.054 | NS |
| SCCG (scr) | −0.429 | NS | −0.418 | NS | −0.316 | NS | −0.359 | NS | 0.119 | NS | −0.169 | NS | −0.015 | NS |
| MRG (%) | 0.205 | NS | 0.217 | NS | 0.108 | NS | 0.169 | NS | 0.198 | NS | −0.038 | NS | 0.029 | NS |
| Genotypic Parameters | FIG (pts) | DPRG (%) | HCRG (%) | CCRG (%) | DCEG (%) | MTRG (%) | RPRG (%) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| r | p | r | p | r | p | r | p | r | p | r | p | r | p | |
| MP305G (lbs) | −0.556 | 0.011 | −0.535 | 0.015 | −0.472 | 0.036 | −0.461 | 0.041 | 0.168 | NS | −0.280 | NS | −0.199 | NS |
| MPFAT305G (lbs) | −0.386 | NS | −0.403 | NS | −0.251 | NS | −0.274 | NS | −0.408 | NS | 0.040 | NS | 0.289 | NS |
| MPPROT305G (lbs) | −0.512 | 0.021 | −0.488 | 0.029 | −0.420 | NS | −0.460 | 0.041 | 0.099 | NS | −0.285 | NS | −0.289 | NS |
| PF305G (%) | 0.130 | NS | 0.100 | 0.029 | 0.160 | 0.029 | 0.150 | NS | −0.427 | NS | 0.248 | NS | 0.380 | NS |
| PP305G (%) | 0.360 | NS | 0.348 | NS | 0.309 | NS | 0.272 | NS | −0.158 | NS | 0.193 | NS | 0.055 | NS |
| SCCG (scr) | −0.211 | NS | −0.203 | NS | −0.076 | NS | −0.300 | NS | 0.192 | NS | 0.188 | NS | −0.569 | 0.009 |
| MRG (%) | 0.555 | 0.011 | 0.546 | 0.013 | 0.437 | NS | 0.488 | 0.029 | 0.208 | NS | 0.164 | NS | 0.184 | NS |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gutiérrez-Reinoso, M.A.; Aponte, P.M.; Cabezas, J.; Rodriguez-Alvarez, L.; Garcia-Herreros, M. Genomic Evaluation of Primiparous High-Producing Dairy Cows: Inbreeding Effects on Genotypic and Phenotypic Production–Reproductive Traits. Animals 2020, 10, 1704. https://doi.org/10.3390/ani10091704
Gutiérrez-Reinoso MA, Aponte PM, Cabezas J, Rodriguez-Alvarez L, Garcia-Herreros M. Genomic Evaluation of Primiparous High-Producing Dairy Cows: Inbreeding Effects on Genotypic and Phenotypic Production–Reproductive Traits. Animals. 2020; 10(9):1704. https://doi.org/10.3390/ani10091704
Chicago/Turabian StyleGutiérrez-Reinoso, Miguel A., Pedro Manuel Aponte, Joel Cabezas, Lleretny Rodriguez-Alvarez, and Manuel Garcia-Herreros. 2020. "Genomic Evaluation of Primiparous High-Producing Dairy Cows: Inbreeding Effects on Genotypic and Phenotypic Production–Reproductive Traits" Animals 10, no. 9: 1704. https://doi.org/10.3390/ani10091704
APA StyleGutiérrez-Reinoso, M. A., Aponte, P. M., Cabezas, J., Rodriguez-Alvarez, L., & Garcia-Herreros, M. (2020). Genomic Evaluation of Primiparous High-Producing Dairy Cows: Inbreeding Effects on Genotypic and Phenotypic Production–Reproductive Traits. Animals, 10(9), 1704. https://doi.org/10.3390/ani10091704







