Lactobacillus reuteri and Enterococcus faecium from Poultry Gut Reduce Mucin Adhesion and Biofilm Formation of Cephalosporin and Fluoroquinolone-Resistant Salmonella enterica
Abstract
:Simple Summary
Abstract
1. Introduction
2. Methodology
2.1. Isolation of Lactic Acid Bacteria
2.2. Test Pathogens
2.3. Safety Assessments
2.3.1. Hemolytic Activity
2.3.2. DNase Activity
2.3.3. Antibiotic Resistance Profiling
2.4. Identification of the Lactic Acid Bacterial Isolates
2.5. Screening of Probiotic Properties of LAB Isolates
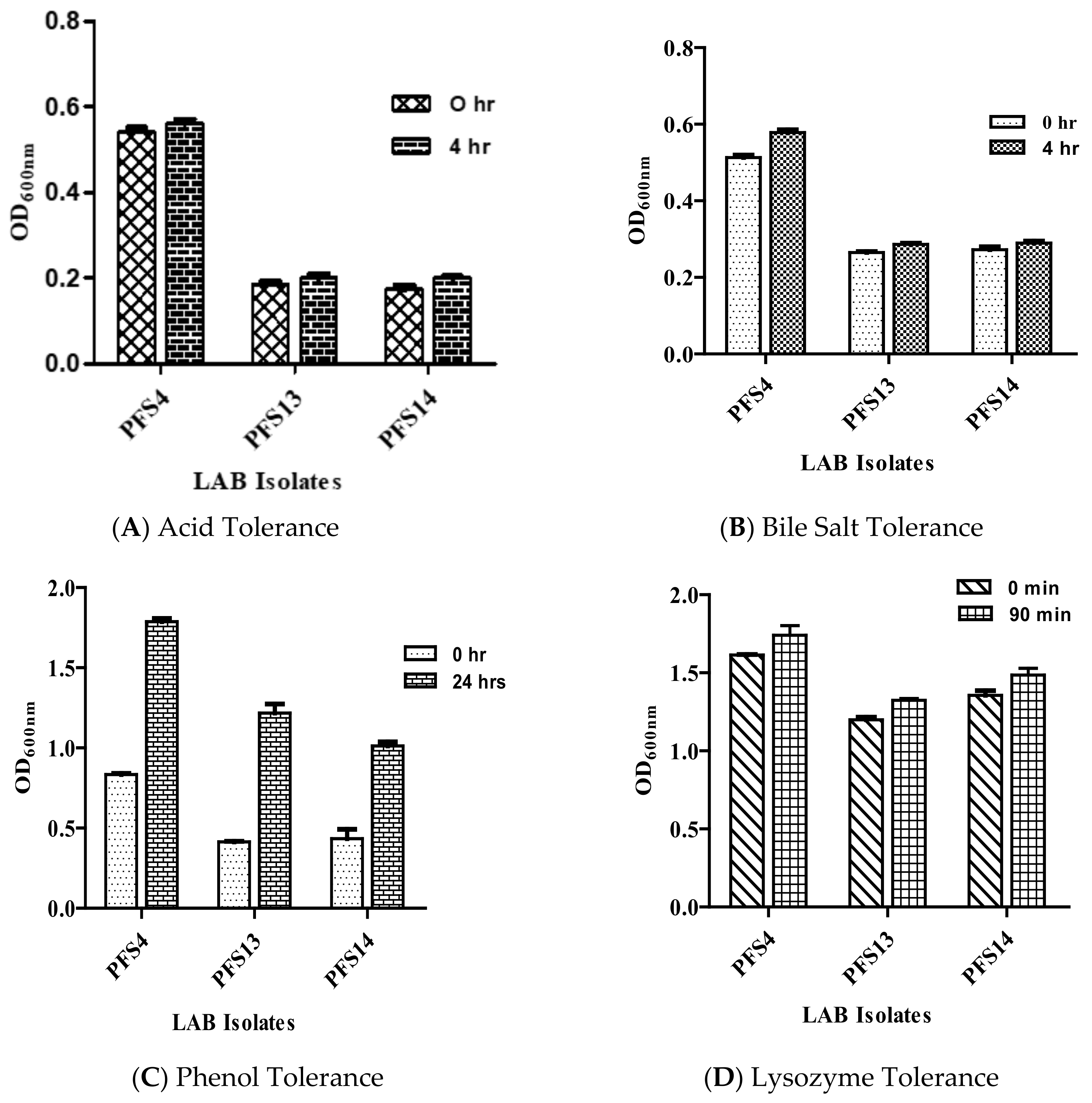
2.5.1. Acid Tolerance
2.5.2. Bile Tolerance
2.5.3. Lysozyme Tolerance
2.5.4. Phenol Tolerance
2.6. Cell Surface Properties
2.6.1. Auto Aggregation and Co-Aggregation Assay
2.6.2. Microbial Adhesion to Hydrocarbon Test (MATH)
2.6.3. Mucin Adhesion Assay
2.6.4. Biofilm Formation Potential of Lactic Acid Bacteria
2.7. Antimicrobial Potential Assessment
2.7.1. Inhibition of Pathogenic Biofilm Formation
2.7.2. Scanning Electron Microscopy
2.7.3. Determination of the Antimicrobial Activity of the LAB Isolates
2.8. Statistical Analysis
3. Results
3.1. Bacterial Isolates
3.2. Safety Assessment
Hemolytic, DNase, and Antibiotic Susceptibility Assay
3.3. Evaluation of Probiotic Properties
Tolerance to GIT Related Stresses
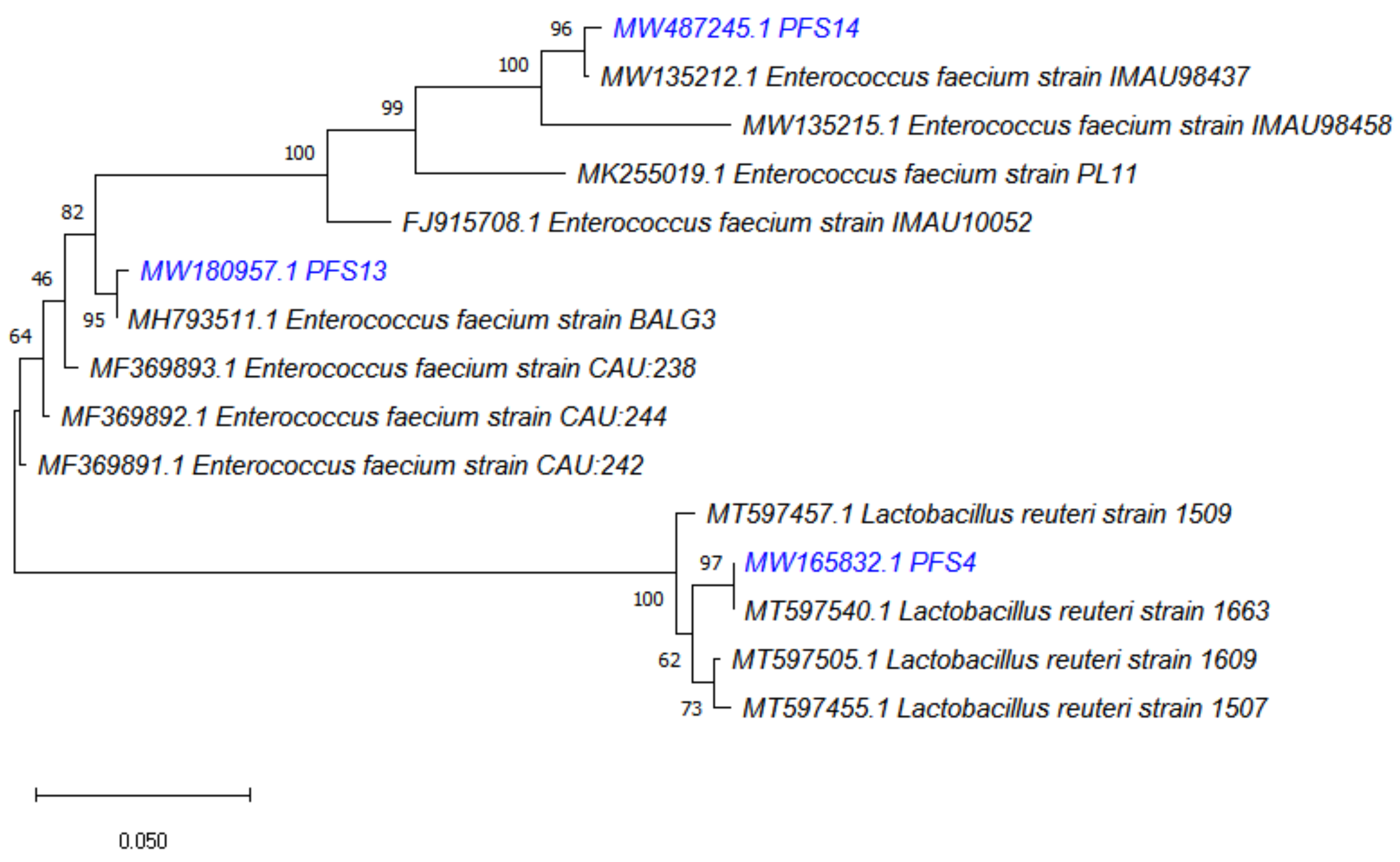
3.4. Molecular Identification of LAB Strains
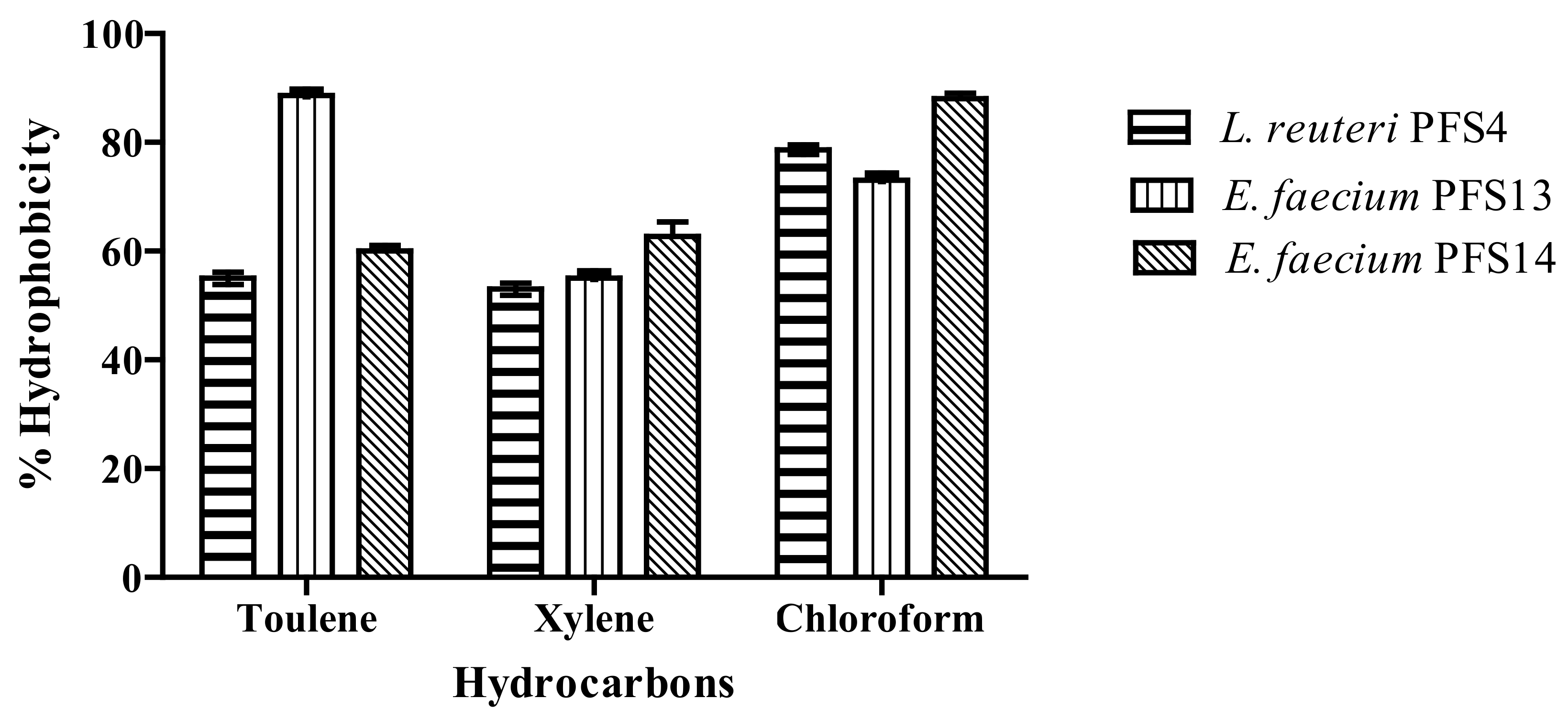
3.5. Cell Surface Hydrophobicity
3.6. Auto-Aggregation and Co-Aggregation Assay
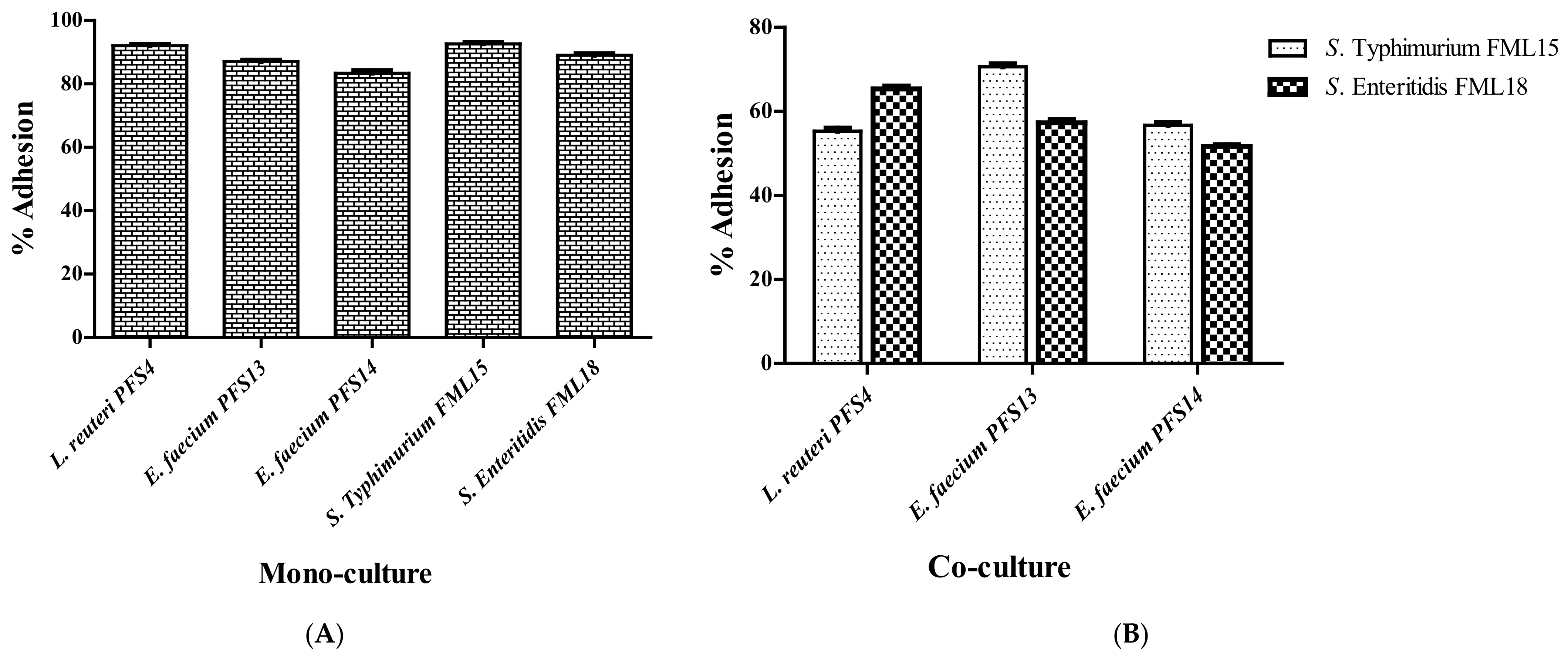
3.7. In Vitro Mucin Adhesion Assay
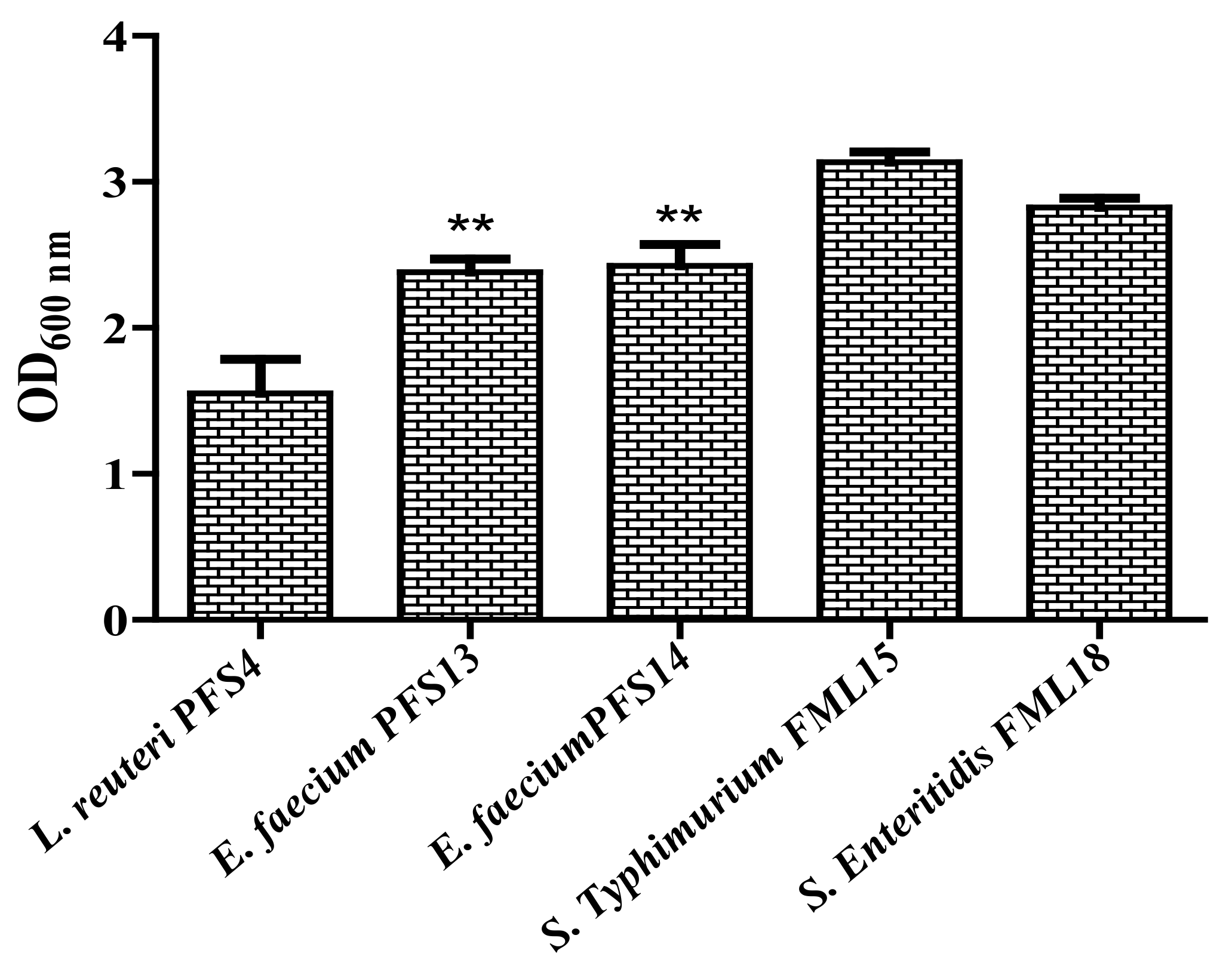
3.8. Biofilm Formation Assay
3.9. Effect of CFS on Biofilm Formation Ability of Salmonella enterica
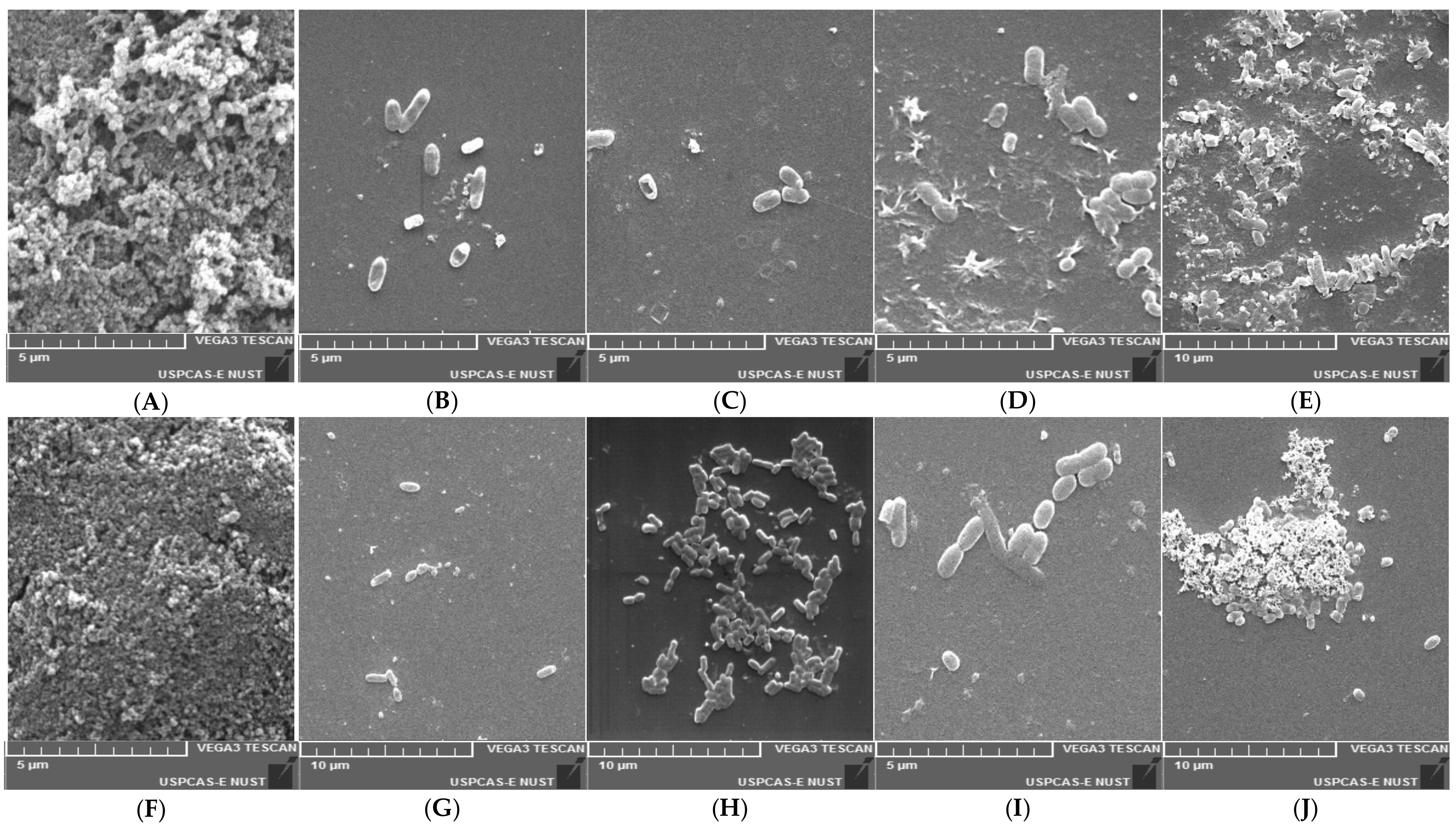
3.10. Scanning Electron Microscopic Study
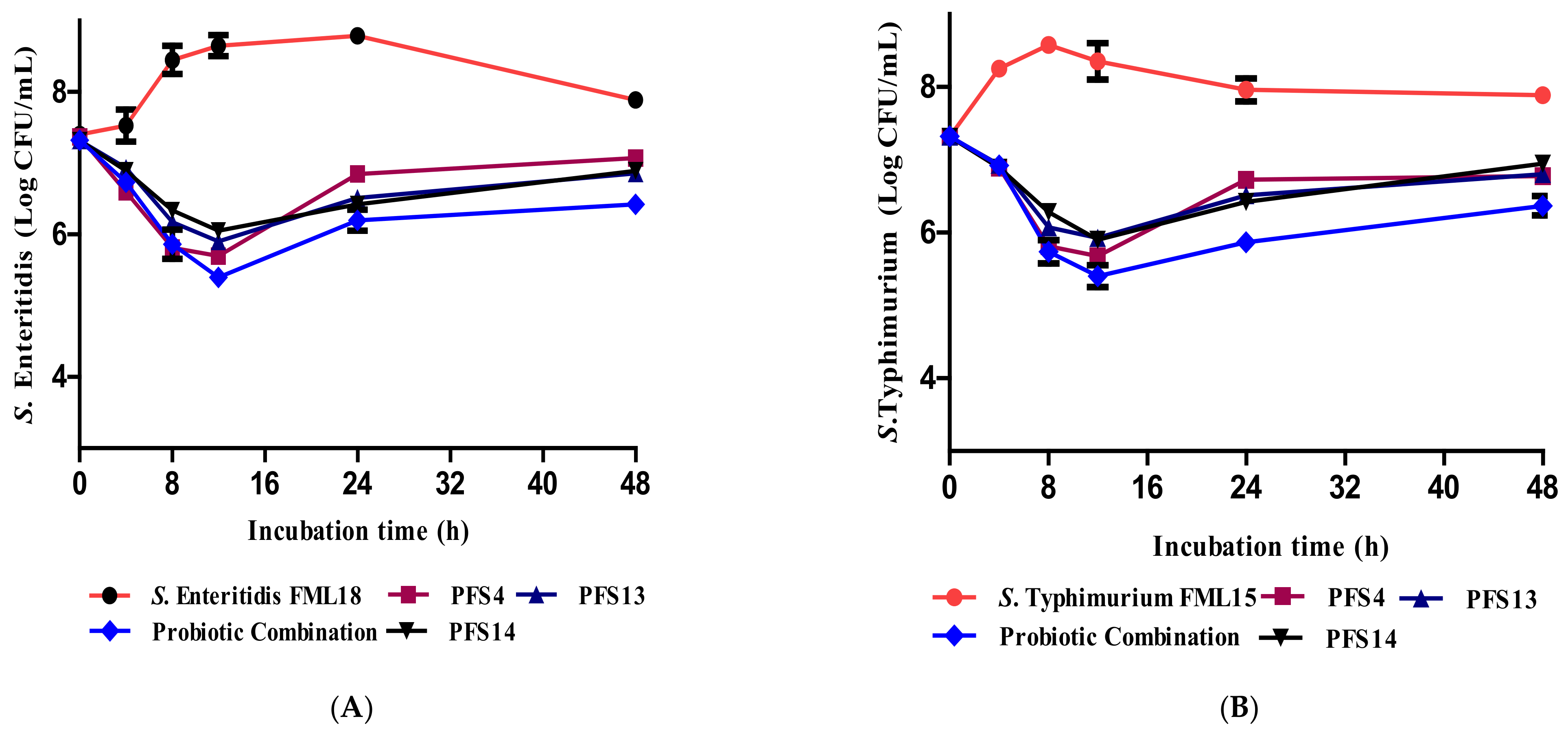
3.11. Effect of CFS on the Growth Kinetics of Salmonella enterica Serovars
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Adetoye, A.; Pinloche, E.; Adeniyi, B.A.; Ayeni, F.A. Characterization and anti-salmonella activities of lactic acid bacteria isolated from cattle faeces. BMC Microbiol. 2018, 18, 96. [Google Scholar] [CrossRef] [Green Version]
- Nhung, N.; Van, N.; Van Cuong, N.; Duong, T.; Nhat, T.; Hang, T.; Nhi, N.; Kiet, B.; Hien, V.; Ngoc, P.; et al. Antimicrobial residues and resistance against critically important antimicrobials in non-typhoidal Salmonella from meat sold at wet markets and supermarkets in Vietnam. Int. J. Food Microbiol. 2018, 266, 301–309. [Google Scholar] [CrossRef] [PubMed]
- Azam, A.; Shafique, M. Agriculture in Pakistan and its Impact on Economy. Int. J. Adv. Sci. Technol. 2017, 103, 47–60. [Google Scholar] [CrossRef]
- Rafique, M.; Potter, R.F.; Ferreiro, A.; Wallace, M.A.; Rahim, A.; Malik, A.A.; Siddique, N.; Abbas, M.A.; D’Souza, A.W.; Burnham, C.; et al. Genomic characterization of antibiotic resistant Escherichia coli isolated from domestic chickens in Pakistan. Front. Microbiol. 2020, 10, 3052. [Google Scholar] [CrossRef]
- Cox, J.; Pavic, A. Advances in enteropathogen control in poultry production. J. Appl. Microbiol. 2010, 108, 745–755. [Google Scholar] [CrossRef] [PubMed]
- Cortés-Sánchez, A.d.J.; Garcia-Barrientos, R.; Minor-Pérez, H.; Dublán-García, O.; San Martin-Azocar, A. Food Safety and Antimicrobial Resistance an Approach to the Genus Salmonella spp. J. Biosci. Med. 2017, 5, 55. [Google Scholar]
- Heredia, N.; García, S. Animals as sources of food-borne pathogens: A review. Anim. Nutr. 2018, 4, 250–255. [Google Scholar] [CrossRef]
- Gut, A.M.; Vasiljevic, T.; Yeager, T.; Donkor, O.N. Salmonella infection–prevention and treatment by antibiotics and probiotic yeasts: A review. Microbiology 2018, 164, 1327–1344. [Google Scholar] [CrossRef] [PubMed]
- Hur, J.; Jawale, C.; Lee, J.H. Antimicrobial resistance of Salmonella isolated from food animals: A review. Food Res. Int. 2012, 45, 819–830. [Google Scholar] [CrossRef]
- Andoh, L.A.; Ahmed, S.; Olsen, J.E.; Obiri-Danso, K.; Newman, M.J.; Opintan, J.A.; Barco, L.; Dalsgaard, A. Prevalence and characterization of Salmonella among humans in Ghana. Trop. Med. Health 2017, 45, 3. [Google Scholar] [CrossRef] [Green Version]
- Breurec, S.; Reynaud, Y.; Frank, T.; Farra, A.; Costilhes, G.; Weill, F.-X.; Le Hello, S. Serotype distribution and antimicrobial resistance of human Salmonella enterica in Bangui, Central African Republic, from 2004 to 2013. PLoS Negl. Trop. Dis. 2019, 13, e0007917. [Google Scholar] [CrossRef] [Green Version]
- EFSA. The European Union Summary Report on Antimicrobial Resistance in zoonotic and indicator bacteria from humans, animals and food in 2017/2018. EFSA J. 2020, 18, 6007. [Google Scholar]
- EFSA. The European Union Summary Report on Antimicrobial Resistance in zoonotic and indicator bacteria from humans, animals and food in 2018/2019. EFSA J. 2021, 19, 6490. [Google Scholar]
- Prestinaci, F.; Pezzotti, P.; Pantosti, A. Antimicrobial resistance: A global multifaceted phenomenon. Pathog. Glob. Health 2015, 109, 309–318. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Ye, K.; Wei, X.; Cao, J.; Xu, X.; Zhou, G. Occurrence, antimicrobial resistance and biofilm formation of Salmonella isolates from a chicken slaughter plant in China. Food Control 2013, 33, 378–384. [Google Scholar] [CrossRef]
- Peruzy, M.F.; Capuano, F.; Proroga, Y.T.R.; Cristiano, D.; Carullo, M.R.; Murru, N. Antimicrobial Susceptibility Testing for Salmonella Serovars Isolated from Food Samples: Five-Year Monitoring (2015–2019). Antibiotics 2020, 9, 365. [Google Scholar] [CrossRef]
- Lunn, A.D.; Fàbrega, A.; Sánchez-Céspedes, J.; Vila, J. Prevalence of mechanisms decreasing quinolone-susceptibility among Salmonella spp. clinical isolates. Int. Microbiol. 2010, 13, 15–20. [Google Scholar] [PubMed]
- Siddiqui, T.R.; Bibi, S.; Mustufa, M.A.; Ayaz, S.M.; Khan, A. High prevalence of typhoidal Salmonella enterica serovars excreting food handlers in Karachi-Pakistan: A probable factor for regional typhoid endemicity. J. Health Popul. Nutr. 2015, 33, 27. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Venter, H.; Henningsen, M.L.; Begg, S.L. Antimicrobial resistance in healthcare, agriculture and the environment: The biochemistry behind the headlines. Essays Biochem. 2017, 61, 1–10. [Google Scholar] [CrossRef]
- Fijan, S.; Frauwallner, A.; Langerholc, T.; Krebs, B.; ter Haar née Younes, J.A.; Heschl, A.; Mičetić Turk, D.; Rogelj, I. Efficacy of using probiotics with antagonistic activity against pathogens of wound infections: An integrative review of literature. BioMed Res. Int. 2019, 2019, 7585486. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hossain, M.I.; Mizan, M.F.R.; Ashrafudoulla, M.; Nahar, S.; Joo, H.; Jahid, I.; Park, S.; Kim, K.; Ha, S. Inhibitory effects of probiotic potential lactic acid bacteria isolated from kimchi against Listeria monocytogenes biofilm on lettuce, stainless-steel surfaces, and MBEC™ biofilm device. LWT 2020, 118, 108864. [Google Scholar] [CrossRef]
- Prabhurajeshwar, C.; Chandrakanth, K. Evaluation of antimicrobial properties and their substances against pathogenic bacteria in-vitro by probiotic Lactobacilli strains isolated from commercial yoghurt. Clin. Nutr. Exp. 2019, 23, 97–115. [Google Scholar] [CrossRef] [Green Version]
- Merino, L.; Trejo, F.M.; De Antoni, G.; Golowczyc, M.A. Lactobacillus strains inhibit biofilm formation of Salmonella sp. isolates from poultry. Food Res. Int. 2019, 123, 258–265. [Google Scholar] [CrossRef]
- Oyewole, O.F.; Maria, C.O.; Tope, P.S.; Funmi, O.O. In vitro Study of Potential Probiotic Lactic Acid Bacteria Isolated from The Gut of Chickens in Abeokuta, Nigeria. Alex. J. Vet. Sci. 2018, 58, 73–84. [Google Scholar] [CrossRef]
- Sanders, M.E.; Akkermans, L.M.; Haller, D.; Hammerman, C.; Heimbach, J.T.; Hörmannsperger, G.; Huys, G. Safety assessment of probiotics for human use. Gut Microbes 2010, 1, 164–185. [Google Scholar] [CrossRef]
- Drago, L.; Gismondo, M.R.; Lombardi, A.; De Haën, C.; Gozzini, L. Inhibition of in vitro growth of enteropathogens by new Lactobacillus isolates of human intestinal origin. FEMS Microbiol. Lett. 1997, 153, 455–463. [Google Scholar] [CrossRef] [PubMed]
- Gharbi, Y.; Fhoula, I.; Ruas-Madiedo, P.; Afef, N.; Boudabous, A.; Gueimonde, M.; Ouzari, H. In-vitro characterization of potentially probiotic Lactobacillus strains isolated from human microbiota: Interaction with pathogenic bacteria and the enteric cell line HT29. Ann. Microbiol. 2019, 69, 61–72. [Google Scholar] [CrossRef]
- Pellegrino, M.S.; Frola, I.D.; Natanael, B.; Gobelli, D.; Nader-Macias, M.E.; Bogni, C. In vitro characterization of lactic acid bacteria isolated from bovine milk as potential probiotic strains to prevent bovine mastitis. Probiotics Antimicrob. Proteins 2019, 11, 74–84. [Google Scholar] [CrossRef] [PubMed]
- Filho, R.A.; Higgins, J.; Higgins, S.; Gaona, G.; Wolfenden, A.; Tellez, G.; Hargis, B. Ability of bacteriophages isolated from different sources to reduce Salmonella enterica serovar Enteritidis in vitro and in vivo. Poult. Sci. 2007, 86, 1904–1909. [Google Scholar] [CrossRef]
- Vinderola, G.; Gueimonde, M.; Gomez-Gallego, C.; Delfederico, L.; Salminen, S. Correlation between in vitro and in vivo assays in selection of probiotics from traditional species of bacteria. Trends Food Sci. Technol. 2017, 68, 83–90. [Google Scholar] [CrossRef]
- Ashraf, R.; Shah, N.P. Selective and differential enumerations of Lactobacillus delbrueckii subsp. bulgaricus, Streptococcus thermophilus, Lactobacillus acidophilus, Lactobacillus casei and Bifidobacterium spp. in yoghurt—A review. Int. J. Food Microbiol. 2011, 149, 194–208. [Google Scholar] [CrossRef] [PubMed]
- Siddique, A.; Azim, S.; Ali, A.; Andleeb, S.; Ahsan, A.; Imran, M.; Rahman, A. Antimicrobial Resistance Profiling of Biofilm Forming Non Typhoidal Salmonella enterica Isolates from Poultry and Its Associated Food Products from Pakistan. Antibiotics 2021, 10, 785. [Google Scholar] [CrossRef]
- Torshizi, M.; Rahimi, S.; Mojgani, N.; Esmaeilkhanian, S.; Grimes, J. Screening of indigenous strains of lactic acid bacteria for development of a probiotic for poultry. Asian-Australas. J. Anim. Sci. 2008, 21, 1495–1500. [Google Scholar] [CrossRef]
- Taheur, F.B.; Kouidhi, B.; Fdhila, K.; Elabed, H.; Slama, R.B.; Mahdouani, K.; Bakhrouf, A.; Chaieb, K. Anti-bacterial and anti-biofilm activity of probiotic bacteria against oral pathogens. Microb. Pathog. 2016, 97, 213–220. [Google Scholar] [CrossRef]
- Singhal, N.; Singh, N.S.; Mohanty, S.; Singh, P.; Virdi, J.S. Evaluation of probiotic characteristics of lactic acid bacteria isolated from two commercial preparations available in Indian market. Indian J. Microbiol. 2019, 59, 112–115. [Google Scholar] [CrossRef]
- Jorgensen, J.H.; Turnidge, J.D. Susceptibility test methods: Dilution and disk diffusion methods. In Manual of Clinical Microbiology, 11th ed.; American Society of Microbiology: Washington, DC, USA, 2015; pp. 1253–1273. [Google Scholar]
- Vlková, E.; Rada, V.; Popelářová, P.; Trojanová, I.; Killer, J. Antimicrobial susceptibility of bifidobacteria isolated from gastrointestinal tract of calves. Livest. Sci. 2006, 105, 253–259. [Google Scholar] [CrossRef]
- Cheng, H.-R.; Jiang, N. Extremely rapid extraction of DNA from bacteria and yeasts. Biotechnol. Lett. 2006, 28, 55–59. [Google Scholar] [CrossRef] [PubMed]
- Turchi, B.; Mancini, S.; Fratini, F.; Pedonese, F.; Nuvoloni, R.; Bertelloni, F.; Ebani, V.V.; Cerri, D. Preliminary evaluation of probiotic potential of Lactobacillus plantarum strains isolated from Italian food products. World J. Microbiol. Biotechnol. 2013, 29, 1913–1922. [Google Scholar] [CrossRef] [PubMed]
- Rajoka, M.S.R.; Hayat, H.; Sarwar, S.; Mehwish, H.; Ahmad, F.; Hussain, N.; Shah, S.; Khurshid, M.; Siddiqu, M.; Shi, J. Isolation and evaluation of probiotic potential of lactic acid bacteria isolated from poultry intestine. Microbiology 2018, 87, 116–126. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Gómez, N.C.; Ramiro, J.M.; Quecan, B.X.; Franco, B.D.d.M. Use of potential probiotic lactic acid bacteria (LAB) biofilms for the control of Listeria monocytogenes, Salmonella typhimurium, and Escherichia coli O157: H7 biofilms formation. Front. Microbiol. 2016, 7, 863. [Google Scholar] [CrossRef] [Green Version]
- Sharma, S.; Kandasamy, S.; Kavitake, D.; Shetty, P.H. Probiotic characterization and antioxidant properties of Weissella confusa KR780676, isolated from an Indian fermented food. LWT 2018, 97, 53–60. [Google Scholar] [CrossRef]
- Nachtigall, C.; Weber, C.; Rothenburger, S.; Jaros, D.; Rohm, H. Test parameters and cell chain length of Streptococcus thermophilus affect the microbial adhesion to hydrocarbons assay: A methodical approach. FEMS Microbiol. Lett. 2019, 366, fnz150. [Google Scholar] [CrossRef]
- Valeriano, V.; Parungao-Balolong, M.; Kang, D.K. In vitro evaluation of the mucin-adhesion ability and probiotic potential of Lactobacillus mucosae LM 1. J. Appl. Microbiol. 2014, 117, 485–497. [Google Scholar] [CrossRef] [PubMed]
- Saxena, S.; Banerjee, G.; Garg, R.; Singh, M. Comparative study of biofilm formation in Pseudomonas aeruginosa isolates from patients of lower respiratory tract infection. J. Clin. Diagn. Res. 2014, 8, DC09. [Google Scholar] [PubMed]
- Kaur, S.; Sharma, P.; Kalia, N.; Singh, J.; Kaur, S. Anti-biofilm Properties of the Fecal Probiotic Lactobacilli Against Vibrio spp. Front. Cell. Infect. Microbiol. 2018, 8, 120. [Google Scholar] [CrossRef] [Green Version]
- Tareb, R.; Bernardeau, M.; Gueguen, M.; Vernoux, J. In vitro characterization of aggregation and adhesion properties of viable and heat-killed forms of two probiotic Lactobacillus strains and interaction with foodborne zoonotic bacteria, especially Campylobacter jejuni. J. Med. Microbiol. 2013, 62, 637–649. [Google Scholar] [CrossRef]
- Das, D.J.; Shankar, A.; Johnson, J.B.; Thomas, S. Critical insights into antibiotic resistance transferability in probiotic Lactobacillus. Nutrition 2020, 69, 110567. [Google Scholar] [CrossRef]
- Ficoseco, C.A.; Mansilla, F.I.; Maldonado, N.C.; Miranda, H.; Nader-Macias, M.E.F.; Vignolo, G.M. Safety and growth optimization of lactic acid bacteria isolated from feedlot cattle for probiotic formula design. Front. Microbiol. 2018, 9, 2220. [Google Scholar] [CrossRef]
- Musikasang, H.; Tani, A.; H-kittikun, A.; Maneerat, S. Probiotic potential of lactic acid bacteria isolated from chicken gastrointestinal digestive tract. World J. Microbiol. Biotechnol. 2009, 25, 1337–1345. [Google Scholar] [CrossRef]
- Hammerum, A.M. Enterococci of animal origin and their significance for public health. Clin. Microbiol. Infect. 2012, 18, 619–625. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Fang, M.; Hu, Y.; Yang, Y.; Yang, M.; Chen, Y. Characterization of the most abundant Lactobacillus species in chicken gastrointestinal tract and potential use as probiotics for genetic engineering. Acta Biochim. Biophys. Sin. 2014, 46, 612–619. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuleaşan, H.; Çakmakçı, M. Effect of reuterin, produced by Lactobacillus reuteri on the surface of sausages to inhibit the growth of Listeria monocytogenes and Salmonella spp. Food/Nahrung 2002, 46, 408–410. [Google Scholar] [CrossRef]
- Shi, S.; Qi, Z.; Sheng, T.; Tu, J.; Shao, Y.; Qi, K. Antagonistic trait of Lactobacillus reuteri S5 against Salmonella enteritidis and assessment of its potential probiotic characteristics. Microb. Pathog. 2019, 137, 103773. [Google Scholar] [CrossRef] [PubMed]
- Audisio, M.C.; Oliver, G.; Apella, M. Protective effect of Enterococcus faecium J96, a potential probiotic strain, on chicks infected with Salmonella pullorum. J. Food Prot. 2000, 63, 1333–1337. [Google Scholar] [CrossRef] [PubMed]
- Reuben, R.C.; Roy, P.C.; Sarkar, S.L.; Alam, R.-U.; Jahid, I. Isolation, characterization, and assessment of lactic acid bacteria toward their selection as poultry probiotics. BMC Microbiol. 2019, 19, 253. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amin, M.; Adams, M.B.; Burke, C.M.; Bolch, C. Isolation and screening of lactic acid bacteria associated with the gastrointestinal tracts of abalone at various life stages for probiotic candidates. Aquac. Rep. 2020, 17, 100378. [Google Scholar] [CrossRef]
- Blajman, J.; Gaziano, C.; Zbrun, M.V.; Soto, L.; Astesana, D.; Berisvil, A.; Scharpen, A.R.; Signorini, M.; Frizzo, L. In vitro and in vivo screening of native lactic acid bacteria toward their selection as a probiotic in broiler chickens. Res. Vet. Sci. 2015, 101, 50–56. [Google Scholar] [CrossRef]
- García-Hernández, Y.; Pérez-Sánchez, T.; Boucourt, R.; Balcázar, J.L.; Nicoli, J.R.; Moreira-Silva, J.; Rodríguez, Z.; Fuertes, H.; Nuñez, O.; Albelo, N.; et al. Isolation, characterization and evaluation of probiotic lactic acid bacteria for potential use in animal production. Res. Vet. Sci. 2016, 108, 125–132. [Google Scholar] [CrossRef] [PubMed]
- Musikasang, H.; Sohsomboon, N.; Tani, A.; Maneerat, S. Bacteriocin-producing lactic acid bacteria as a probiotic potential from Thai indigenous chickens. Czech J. Anim. Sci. 2012, 57, 137–149. [Google Scholar] [CrossRef] [Green Version]
- Dowarah, R.; Verma, A.; Agarwal, N. The use of Lactobacillus as an alternative of antibiotic growth promoters in pigs: A review. Anim. Nutr. 2017, 3, 1–6. [Google Scholar] [CrossRef]
- Bernardeau, M.; Vernoux, J.P.; Gueguen, M. Safety and efficacy of probiotic lactobacilli in promoting growth in post-weaning Swiss mice. Int. J. Food Microbiol. 2002, 77, 19–27. [Google Scholar] [CrossRef]
- Anadón, A.; Martínez-Larrañaga, M.R.; Martínez, M. Probiotics for animal nutrition in the European Union. Regulation and safety assessment. Regul. Toxicol. Pharmacol. 2006, 45, 91–95. [Google Scholar] [CrossRef]
- Cao, G.; Zeng, X.; Chen, A.; Zhou, L.; Zhang, L.; Xiao, Y.; Yang, C. Effects of a probiotic, Enterococcus faecium, on growth performance, intestinal morphology, immune response, and cecal microflora in broiler chickens challenged with Escherichia coli K88. Poult. Sci. 2013, 92, 2949–2955. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Jeong, J.; Lee, S.; Kim, I. Effect of dietary supplementation with a probiotic (Enterococcus faecium) on production performance, excreta microflora, ammonia emission, and nutrient utilization in ISA brown laying hens. Poult. Sci. 2016, 95, 2829–2835. [Google Scholar] [CrossRef] [PubMed]
- Babot, J.D.; Argañaraz-Martínez, E.; Saavedra, L.; Apella, M.C.; Chaia, A.P. Compatibility and safety of five lectin-binding putative probiotic strains for the development of a multi-strain protective culture for poultry. Benef. Microbes 2018, 9, 927–935. [Google Scholar] [CrossRef]
- Huys, G.; D’HAENE, K.; Danielsen, M.; Maettoe, J.; Egervärn, M.; Vandamme, P. Phenotypic and molecular assessment of antimicrobial resistance in Lactobacillus paracasei strains of food origin. J. Food Prot. 2008, 71, 339–344. [Google Scholar] [CrossRef] [PubMed]
- Sornplang, P.; Soikum, C. Effect of Lactobacillus spp. Supplementation on Spoilage Bacteria Decontamination in Chicken Meat. In Proceedings of the International Conference on Beneficial Microbes 2016, Phuket, Thailand, 31 May–2 June 2016. [Google Scholar]
- Toomey, N.; Bolton, D.; Fanning, S. Characterisation and transferability of antibiotic resistance genes from lactic acid bacteria isolated from Irish pork and beef abattoirs. Res. Microbiol. 2010, 161, 127–135. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.H.; Manuzon, M.; Lehman, M.; Wan, K.; Luo, H.; Wittum, T.E.; Yousef, A.; Bakaletz, L. Food commensal microbes as a potentially important avenue in transmitting antibiotic resistance genes. FEMS Microbiol. Lett. 2006, 254, 226–231. [Google Scholar] [CrossRef] [PubMed]
- Ouoba, L.I.I.; Lei, V.; Jensen, L.B. Resistance of potential probiotic lactic acid bacteria and bifidobacteria of African and European origin to antimicrobials: Determination and transferability of the resistance genes to other bacteria. Int. J. Food Microbiol. 2008, 121, 217–224. [Google Scholar] [CrossRef]
- Pan, L.; Hu, X.; Wang, X. Assessment of antibiotic resistance of lactic acid bacteria in Chinese fermented foods. Food Control 2011, 22, 1316–1321. [Google Scholar] [CrossRef]
- Ashraf, R.; Shah, N.P. Antibiotic resistance of probiotic organisms and safety of probiotic dairy products. Int. Food Res. J. 2011, 18, 837–853. [Google Scholar]
- Giraffa, G. Selection and design of lactic acid bacteria probiotic cultures. Eng. Life Sci. 2012, 12, 391–398. [Google Scholar] [CrossRef]
- Harzallah, D.; Belhadj, H. Lactic acid bacteria as probiotics: Characteristics, selection criteria and role in immunomodulation of human GI muccosal barrier. In Lactic Acid Bacteria—R & D for Food, Health and Livestock Purposes; Kongo, M., Ed.; InTech: London, UK, 2013; pp. 197–216. [Google Scholar]
- Dicks, L.M.; Todorov, S.D.; Franco, B. Current Status of Antibiotic Resistance in Lactic Acid Bacteria; Nova Publisher: New York, NY, USA, 2011; pp. 379–425. [Google Scholar]
- Gueimonde, M.; Sánchez, B.; de Los Reyes-Gavilán, C.G.; Margolles, A. Antibiotic resistance in probiotic bacteria. Front. Microbiol. 2013, 4, 202. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pringsulaka, O.; Rueangyotchanthana, K.; Suwannasai, N.; Watanapokasin, R.; Amnueysit, P.; Sunthornthummas, S.; Sukkhum, S.; Sarawaneeyaruk, S.; Rangsiruji, A. In vitro screening of lactic acid bacteria for multi-strain probiotics. Livest. Sci. 2015, 174, 66–73. [Google Scholar] [CrossRef]
- Pedersen, K.; Tannock, G. Colonization of the porcine gastrointestinal tract by lactobacilli. Appl. Environ. Microbiol. 1989, 55, 279–283. [Google Scholar] [CrossRef] [Green Version]
- Han, Q.; Kong, B.; Chen, Q.; Sun, F.; Zhang, H. In vitro comparison of probiotic properties of lactic acid bacteria isolated from Harbin dry sausages and selected probiotics. J. Funct. Foods 2017, 32, 391–400. [Google Scholar] [CrossRef]
- Ehrmann, M.A.; Kurzak, P.; Bauer, J.; Vogel, R.F. Characterization of lactobacilli towards their use as probiotic adjuncts in poultry. J. Appl. Microbiol. 2002, 92, 966–975. [Google Scholar] [CrossRef]
- Grajek, K.; Sip, A.; Foksowicz-Flaczyk, J.; Dobrowolska, A.; Wita, A. Adhesive and hydrophobic properties of the selected LAB isolated from gastrointestinal tract of farming animals. Acta Biochim. Pol. 2016, 63, 311–314. [Google Scholar] [CrossRef] [Green Version]
- Sharma, K.; Attri, S.; Goel, G. Selection and evaluation of probiotic and functional characteristics of autochthonous lactic acid bacteria isolated from fermented wheat flour dough babroo. Probiotics Antimicrob. Proteins 2019, 11, 774–784. [Google Scholar] [CrossRef]
- Lukic, J.; Strahinic, I.; Milenkovic, M.; Nikolic, M.; Tolinacki, M.; Kojic, M.; Begovic, J. Aggregation factor as an inhibitor of bacterial binding to gut mucosa. Microb. Ecol. 2014, 68, 633–644. [Google Scholar] [CrossRef]
- Klopper, K.B.; Deane, S.M.; Dicks, L.M. Aciduric strains of Lactobacillus reuteri and Lactobacillus rhamnosus, isolated from human feces, have strong adhesion and aggregation properties. Probiotics Antimicrob. Proteins 2018, 10, 89–97. [Google Scholar] [CrossRef] [PubMed]
- Veljović, K.; Popović, N.; Miljković, M.; Tolinački, M.; Terzić-Vidojević, A.; Kojić, M. Novel aggregation promoting factor AggE contributes to the probiotic properties of Enterococcus faecium BGGO9-28. Front. Microbiol. 2017, 8, 1843. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chaucheyras-Durand, F.; Durand, H. Probiotics in animal nutrition and health. Benef. Microbes 2010, 1, 3–9. [Google Scholar] [CrossRef] [PubMed]
- Dhama, K.; Verma, V.; Sawant, P.; Tiwari, R.; Vaid, R.; Chauhan, R. Applications of probiotics in poultry: Enhancing immunity and beneficial effects on production performances and health: A review. J. Immunol. Immunopathol. 2011, 13, 1–19. [Google Scholar]
- Zhang, W.; Liu, M.; Dai, X. Biological characteristics and probiotic effect of Leuconostoc lactis strain isolated from the intestine of black porgy fish. Braz. J. Microbiol. 2013, 44, 685–691. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- TAS, G.I.A.-C.; Korany, A.; Bustos, I.; de Cadi, N. Anos LPGO, Requena T, Pel A Ez C, Mart I Nez-Cuesta MC. Adhesion abilities of dairy Lactobacillus plantarum strains showing an aggregation phenotype. Food Res. Int. 2014, 57, 44–50. [Google Scholar]
- Saulnier, D.M.; Ringel, Y.; Heyman, M.B.; Foster, J.A.; Bercik, P.; Shulman, R.J.; Versalovic, J.; Verdu, E.F.; Dinan, T.G.; Hecht, G.; et al. The intestinal microbiome, probiotics and prebiotics in neurogastroenterology. Gut Microbes 2013, 4, 17–27. [Google Scholar] [CrossRef]
- Li, X.; Yue, L.; Guan, X.; Qiao, S. The adhesion of putative probiotic lactobacilli to cultured epithelial cells and porcine intestinal mucus. J. Appl. Microbiol. 2008, 104, 1082–1091. [Google Scholar] [CrossRef]
- Martens, E.C.; Neumann, M.; Desai, M.S. Interactions of commensal and pathogenic microorganisms with the intestinal mucosal barrier. Nat. Rev. Microbiol. 2018, 16, 457–470. [Google Scholar] [CrossRef]
- Uraipan, S.; Hongpattarakere, T. Antagonistic characteristics against food-borne pathogenic bacteria of lactic acid bacteria and bifidobacteria isolated from feces of healthy Thai infants. Jundishapur J. Microbiol. 2015, 8, e18264. [Google Scholar] [CrossRef] [Green Version]
- Jin, L.; Marquardt, R.; Zhao, X. A strain of Enterococcus faecium (18C23) inhibits adhesion of enterotoxigenic Escherichia coli K88 to porcine small intestine mucus. Appl. Environ. Microbiol. 2000, 66, 4200–4204. [Google Scholar] [CrossRef] [Green Version]
- Jonsson, H.; Ström, E.; Roos, S. Addition of mucin to the growth medium triggers mucus-binding activity in different strains of Lactobacillus reuteri in vitro. FEMS Microbiol. Lett. 2001, 204, 19–22. [Google Scholar] [CrossRef] [Green Version]
- Ventolini, G.; Mitchell, E.; Salazar, M. Biofilm formation by vaginal Lactobacillus in vivo. Med. Hypotheses 2015, 84, 417–420. [Google Scholar] [CrossRef] [PubMed]
- Terraf, M.L.; Tomás, M.J.; Nader-Macías, M.; Silva, C. Screening of biofilm formation by beneficial vaginal lactobacilli and influence of culture media components. J. Appl. Microbiol. 2012, 113, 1517–1529. [Google Scholar] [CrossRef] [PubMed]
- Dec, M.; Nowaczek, A.; Urban-Chmiel, R.; Stępień-Pyśniak, D.; Wernicki, A. Probiotic potential of Lactobacillus isolates of chicken origin with anti-Campylobacter activity. J. Vet. Med. Sci. 2018, 80, 1195–1203. [Google Scholar] [CrossRef] [Green Version]
- Balakrishna, A. In vitro evaluation of adhesion and aggregation abilities of four potential probiotic strains isolated from guppy (Poecilia reticulata). Braz. Arch. Biol. Technol. 2013, 56, 793–800. [Google Scholar] [CrossRef] [Green Version]
- Nakphaichit, M.; Sobanbua, S.; Siemuang, S.; Vongsangnak, W.; Nakayama, J.; Nitisinprasert, S. Protective effect of Lactobacillus reuteri KUB-AC5 against Salmonella Enteritidis challenge in chickens. Benef. Microbes 2019, 10, 43–54. [Google Scholar] [CrossRef] [PubMed]
- Arena, M.P.; Capozzi, V.; Spano, G.; Fiocco, D. The potential of lactic acid bacteria to colonize biotic and abiotic surfaces and the investigation of their interactions and mechanisms. Appl. Microbiol. Biotechnol. 2017, 101, 2641–2657. [Google Scholar] [CrossRef]
- Camargo, A.C.; Todorov, S.D.; Chihib, N.E.; Drider, D.; Nero, L.A. Lactic acid bacteria (LAB) and their bacteriocins as alternative biotechnological tools to control Listeria monocytogenes biofilms in food processing facilities. Mol. Biotechnol. 2018, 60, 712–726. [Google Scholar] [CrossRef]
- Bjarnsholt, T. The role of bacterial biofilms in chronic infections. Apmis 2013, 121, 1–58. [Google Scholar] [CrossRef] [PubMed]
- Janssens, J.C.; Steenackers, H.; Robijns, S.; Gellens, E.; Levin, J.; Zhao, H.; Hermans, K.; De Coster, D.; Verhoeven, T.L.; Marchal, K.; et al. Brominated furanones inhibit biofilm formation by Salmonella enterica serovar Typhimurium. Appl. Environ. Microbiol. 2008, 74, 6639–6648. [Google Scholar] [CrossRef] [Green Version]
- Kecerová, K.; Pristaš, P.; Javorský, P. Bacteriocin production and sensitivity. Folia Microbiol. 2004, 49, 172. [Google Scholar] [CrossRef] [PubMed]
- Lima, E.T.; Filho, R.L.A.; Okamoto, A.S.; Noujaim, J.C.; Barros, M.R.; Crocci, A. Evaluation in vitro of the antagonistic substances produced by Lactobacillus spp. isolated from chickens. Can. J. Vet. Res. 2007, 71, 103–107. [Google Scholar]
- Martin, R.; Olivares, M.; Marin, M.; Xaus, J.; Fernández, L.; Rodríguez, J. Characterization of a reuterin-producing Lactobacillus coryniformis strain isolated from a goat’s milk cheese. Int. J. Food Microbiol. 2005, 104, 267–277. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.-C.; Lai, C.-C.; Huang, H.-L.; Huang, W.-Y.; Toh, H.-S.; Weng, T.-C.; Chuang, Y.-C.; Lu, Y.-C.; Tang, H.-J. Antimicrobial activity of Lactobacillus species against carbapenem-resistant Enterobacteriaceae. Front. Microbiol. 2019, 10, 789. [Google Scholar] [CrossRef] [PubMed]
- El-Mokhtar, M.A.; Hassanein, K.M.; Ahmed, A.S.; Gad, G.F.; Amin, M.M.; Hassanein, O. Antagonistic activities of cell-free supernatants of lactobacilli against extended-spectrum β-lactamase producing Klebsiella pneumoniae and Pseudomonas aeruginosa. Infect. Drug Resist. 2020, 13, 543–552. [Google Scholar] [CrossRef] [Green Version]

| Isolates | CN | CIP | K | VA | C | STM | IPM | MEM | NA | CFM | FEP | LZD | SXT | RIF | AK | TE | AMC |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PFS1 | R | R | R | R | S | R | S | S | S | R | R | S | R | S | R | R | S |
| PFS2 | R | R | R | R | S | R | S | S | S | R | R | S | S | S | R | S | S |
| PFS3 | R | S | R | R | R | R | S | I | S | R | R | S | R | S | R | S | I |
| PFS4 | S | S | R | R | S | R | S | S | S | S | S | S | S | S | S | S | S |
| PFS5 | R | S | R | R | R | R | S | S | S | R | R | S | R | S | R | R | S |
| PFS6 | R | R | R | R | S | S | S | S | S | S | R | S | R | S | S | S | I |
| PFS7 | R | S | R | R | R | R | S | S | S | R | R | S | R | S | R | R | I |
| PFS8 | R | R | R | R | S | R | S | S | S | R | R | S | I | S | R | S | S |
| PFS9 | R | S | R | R | I | R | S | S | S | R | I | S | I | S | R | R | R |
| PFS10 | R | S | R | S | S | R | S | S | S | R | I | S | I | S | R | S | I |
| PFS11 | S | S | R | S | S | R | S | S | S | R | R | S | R | S | R | S | R |
| PFS12 | R | S | R | S | R | S | S | S | S | R | R | S | I | S | R | S | S |
| PFS13 | S | S | R | S | S | S | S | S | S | S | S | S | S | S | S | R | S |
| PFS14 | S | S | R | S | S | R | S | S | S | S | S | S | S | S | S | S | S |
| PFS15 | S | R | R | R | S | R | S | S | S | R | R | S | R | S | R | R | S |
| PFS16 | S | S | R | R | S | R | S | R | S | R | I | S | I | S | R | I | S |
| PFS17 | S | S | R | S | S | R | S | I | S | R | R | S | I | S | R | I | S |
| Auto-Aggregation (%) | Co-Aggregation (%) | ||
|---|---|---|---|
| Strains | S. Typhimurium FML15 | S. Enteritidis FML18 | |
| L. reuteri PFS4 | 80.1 ± 3.1 * | 39.3 ± 4.1 | 42.1 ± 1.1 * |
| E. faecium PF13 | 71.2 ± 2.1 | 66.5 ± 1.1 * | 71.4 ± 1.5 * |
| E. faecium PF14 | 76.2 ± 1.8 * | 40.1 ± 1.8 * | 43.5 ± 1.1 |
| Probiotic combination | 61.1 ± 0.8 | 52.2 ± 2.3 * | 68.6 ± 1.1 * |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Siddique, A.; Azim, S.; Ali, A.; Adnan, F.; Arif, M.; Imran, M.; Ganda, E.; Rahman, A. Lactobacillus reuteri and Enterococcus faecium from Poultry Gut Reduce Mucin Adhesion and Biofilm Formation of Cephalosporin and Fluoroquinolone-Resistant Salmonella enterica. Animals 2021, 11, 3435. https://doi.org/10.3390/ani11123435
Siddique A, Azim S, Ali A, Adnan F, Arif M, Imran M, Ganda E, Rahman A. Lactobacillus reuteri and Enterococcus faecium from Poultry Gut Reduce Mucin Adhesion and Biofilm Formation of Cephalosporin and Fluoroquinolone-Resistant Salmonella enterica. Animals. 2021; 11(12):3435. https://doi.org/10.3390/ani11123435
Chicago/Turabian StyleSiddique, Abubakar, Sara Azim, Amjad Ali, Fazal Adnan, Maryum Arif, Muhammad Imran, Erika Ganda, and Abdur Rahman. 2021. "Lactobacillus reuteri and Enterococcus faecium from Poultry Gut Reduce Mucin Adhesion and Biofilm Formation of Cephalosporin and Fluoroquinolone-Resistant Salmonella enterica" Animals 11, no. 12: 3435. https://doi.org/10.3390/ani11123435
APA StyleSiddique, A., Azim, S., Ali, A., Adnan, F., Arif, M., Imran, M., Ganda, E., & Rahman, A. (2021). Lactobacillus reuteri and Enterococcus faecium from Poultry Gut Reduce Mucin Adhesion and Biofilm Formation of Cephalosporin and Fluoroquinolone-Resistant Salmonella enterica. Animals, 11(12), 3435. https://doi.org/10.3390/ani11123435








