Isolation, Identification and Characterization of a Novel Megalocytivirus from Cultured Tilapia (Oreochromis spp.) from Southern California, USA
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Clinical History
2.2. Bacterial Isolation and Identification
2.3. Virus Isolation
2.4. Transmission Electron Microscopy
2.5. Virus Identification by PCR
2.6. Next Generation Sequencing and Phylogenetic Analysis
2.7. Quantitative PCR for Detection of the Novel Megalocytivirus
2.8. Experimental Challenge
2.9. Histopathology
2.10. Statistical Analysis
3. Results
3.1. Bacterial Isolation and Identification from Naturally Infected Fish
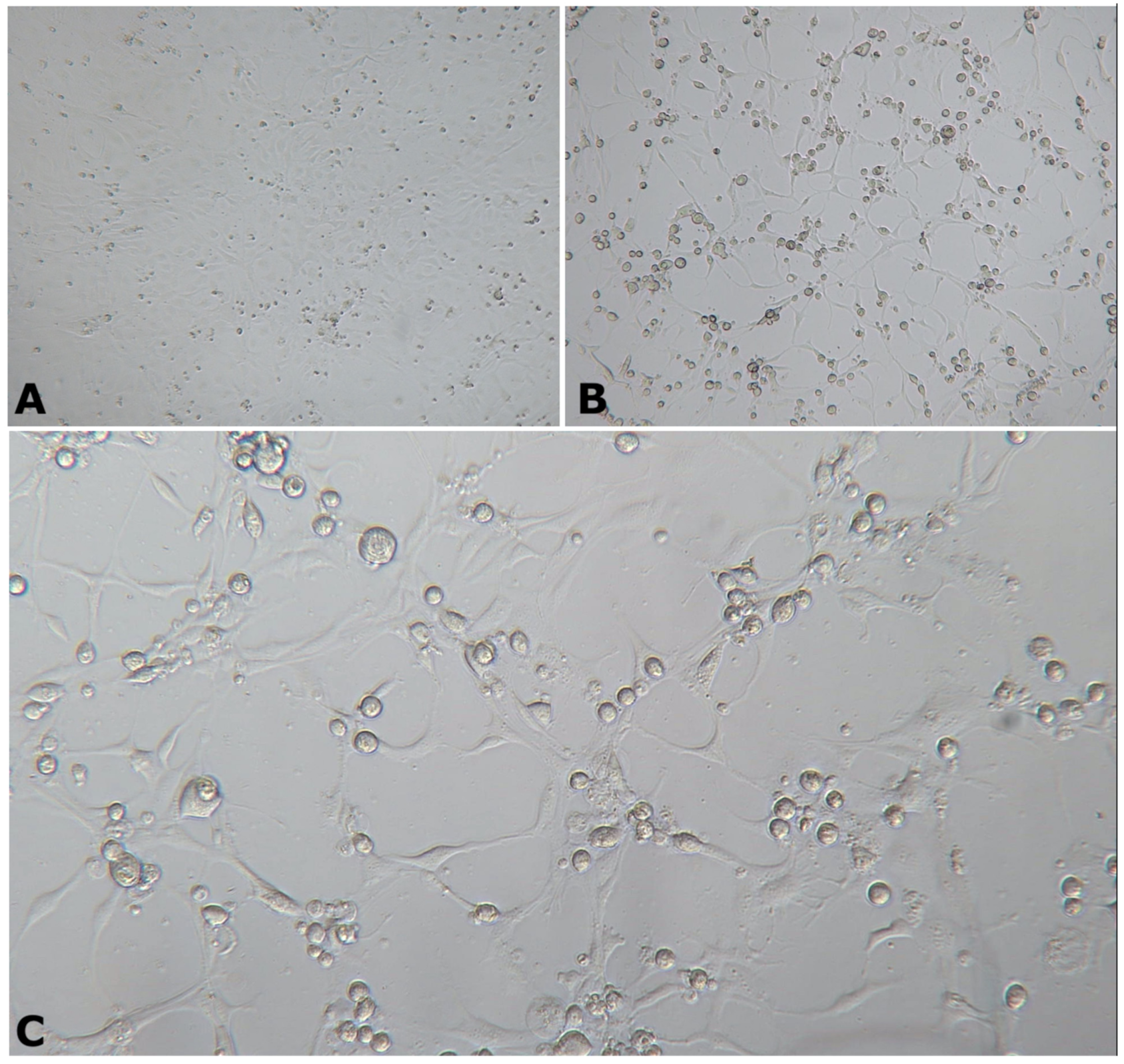
3.2. Viral Identification and Characterization from Naturally Infected Fish
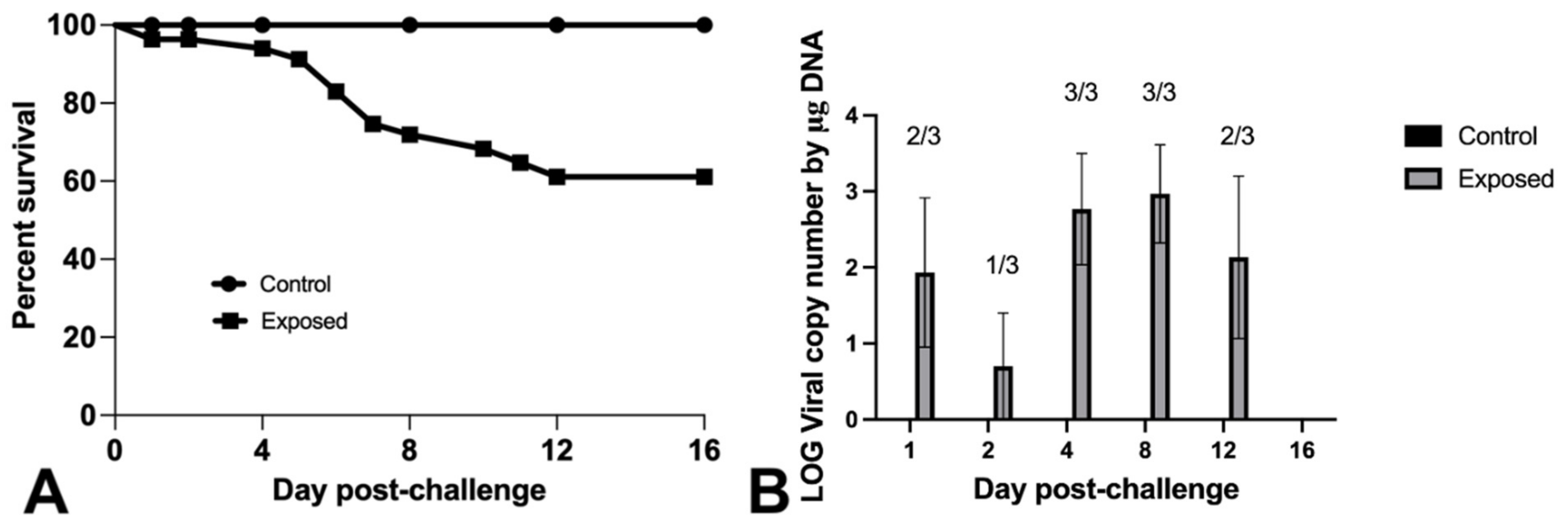
3.3. Experimental Challenge
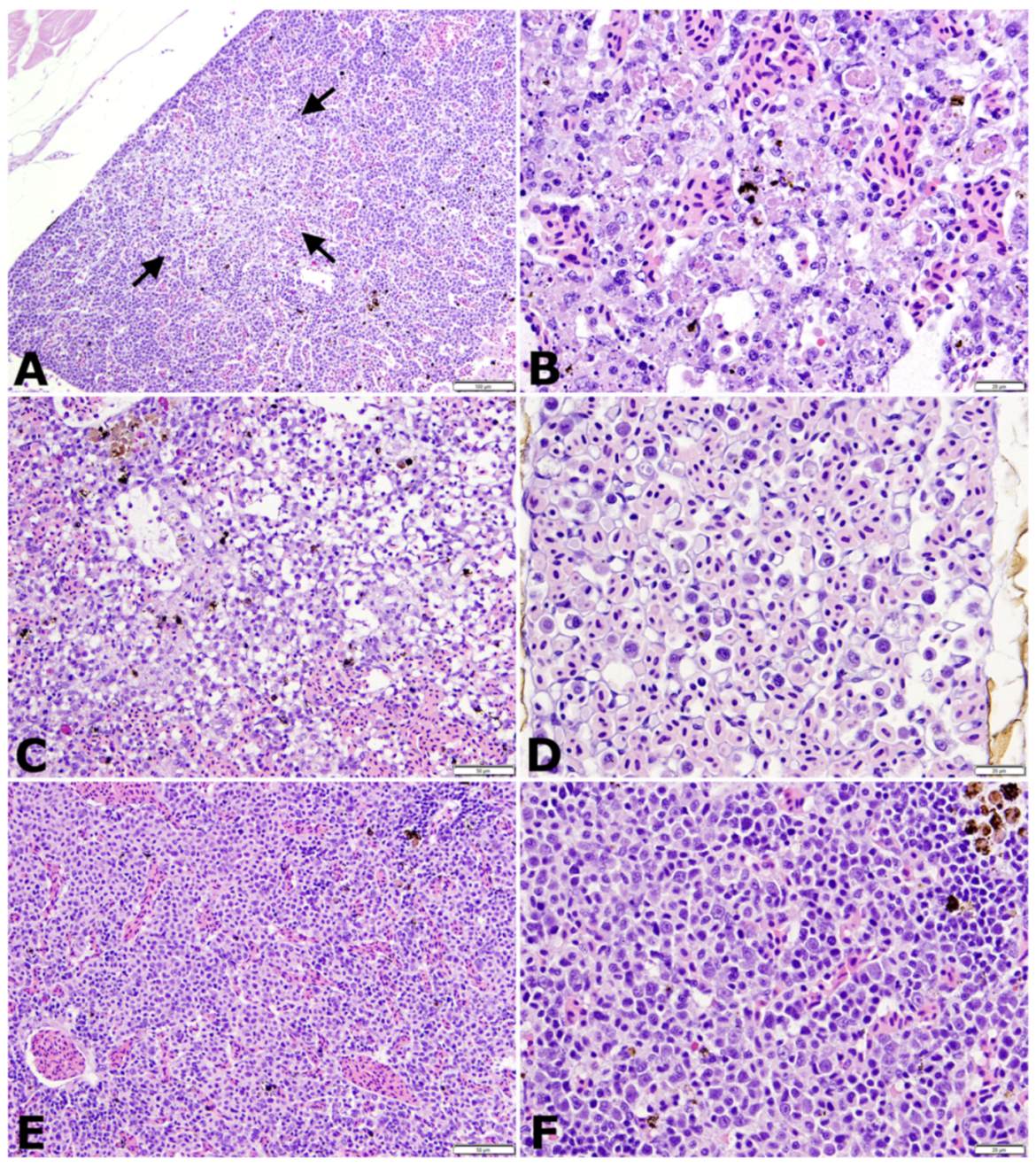
3.4. Histopathology
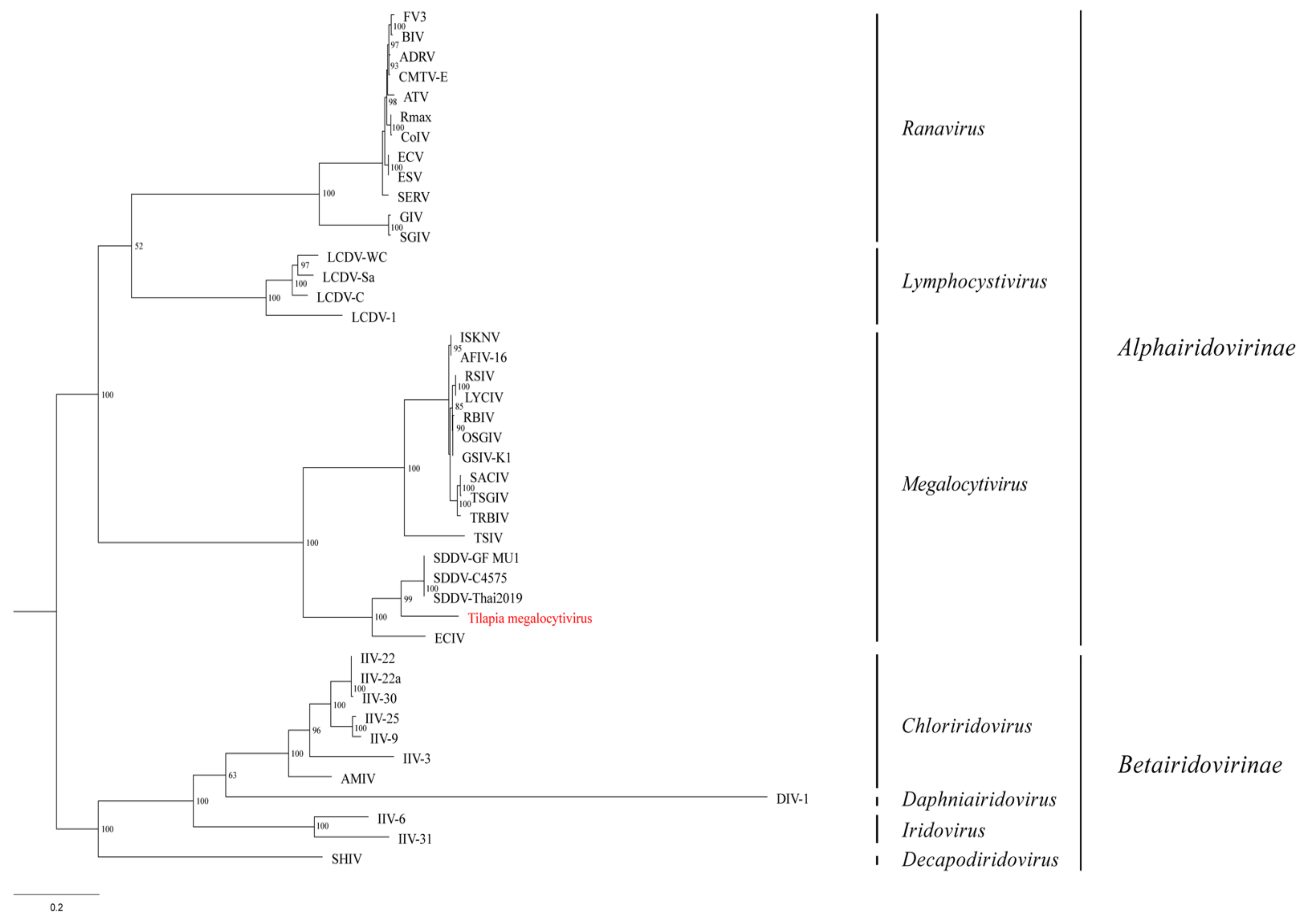
3.5. Phylogenetic Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Food and Agriculture Organization of the United Nations (FAO). Cultured Aquatic Species Information Program Oreochromis niloticus (Linnaeus, 1758). Available online: http://www.fao.org/fishery/culturedspecies/Oreochromis_niloticus/en (accessed on 31 March 2021).
- Shelton, W.L.; Popma, T.J. Biology. In Tilapia: Biology, Culture, and Nutrition, 1st ed.; Lim, C.E., Webster, C.D., Eds.; Taylor and Francis: Abingdon, UK, 2006; pp. 1–49. [Google Scholar]
- Surachetpong, W.; Roy, S.R.K.; Nicholson, P. Tilapia Lake virus: The story so far. J. Fish. Dis. 2020, 43, 1115–1132. [Google Scholar] [CrossRef]
- Liu, W.; Zhang, Y.; Ma, J.; Jiang, N.; Fan, Y.; Zhou, Y.; Cain, K.; Yi, M.; Jia, K.; Wen, H.; et al. Determination of a novel parvovirus pathogen associated with massive mortality in adult tilapia. PLoS Pathog. 2020, 16, e1008765. [Google Scholar] [CrossRef] [PubMed]
- Subramaniam, K.; Gotesman, M.; Smith, C.E.; Steckler, N.K.; Kelley, K.L.; Groff, J.M.; Waltzek, T.B. Megalocytivirus infection in cultured Nile tilapia, Oreochromis niloticus. Dis. Aquat. Organ. 2016, 119, 253–258. [Google Scholar] [CrossRef]
- Ramirez-Paredes, J.G.; Paley, R.K.; Hunt, W.; Feist, S.W.; Stone, D.M.; Field, T.; Haydon, D.J.; Ziddah, P.A.; Duodu, S.; Wallis, T.S.; et al. First detection of Infectious Spleen and kidney Necrosis Virus (ISKNV) associated with massive mortalities in farmed tilapia in Africa. Transbound. Emerg. Dis. 2021, 68, 1550–1563. [Google Scholar] [CrossRef] [PubMed]
- Kurita, J.; Nakajima, K. Megalocytiviruses. Viruses 2012, 4, 521–538. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chinchar, V.G.; Hick, P.; Ince, I.A.; Jancovich, J.K.; Marschang, R.; Qin, Q.; Subramaniam, K.; Waltzek, T.B.; Whittington, R.; Williams, T.; et al. ICTV Virus Taxonomy Profile: Iridoviridae. J. Gen. Virol. 2017, 98, 890–891. [Google Scholar] [CrossRef]
- Sriwanayos, P.; Francis-Floyd, R.; Stidworthy, M.F.; Petty, B.D.; Kelley, K.; Waltzek, T.B. Megalocytivirus infection in orbiculate batfish Platax orbicularis. Dis. Aquat. Organ. 2013, 105, 1–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Inouye, K.; Yamano, K.; Maeno, Y.; Nakajima, K.; Matsuoka, M.; Wada, Y.; Sorinachi, M. Iridovirus infection of cultured red-sea bream, Pagrus major. Fish Pathol. 1992, 27, 19–27. [Google Scholar] [CrossRef]
- Kim, K.I.; Hwang, S.D.; Cho, M.Y.; Jung, S.H.; Kim, Y.C.; Jeong, H.D. A natural infection by the red sea bream iridovirus-type Megalocytivirus in the golden mandarin fish Siniperca scherzeri. J. Fish. Dis. 2018, 41, 1229–1233. [Google Scholar] [CrossRef] [PubMed]
- Whittington, R.J.; Becker, J.A.; Dennis, M.M. Irodovirus infections in finfish—Critical review with emphasis on ranaviruses. J. Fish. Dis. 2010, 33, 95–122. [Google Scholar] [CrossRef] [PubMed]
- Hick, P.; Becker, J.; Whittington, R. Chapter 8—Iridoviruses of fish. In Aquaculture Virology; Kibenge, F.S.B., Godoy, M.G., Eds.; Academic Press: San Diego, CA, USA, 2016; pp. 127–152. [Google Scholar]
- Tanaka, N.; Izawa, T.; Kuwamura, M.; Higashiguchi, N.; Kezuka, C.; Kurata, O.; Wada, S.; Yamate, J. The first case of infectious spleen and kidney necrosis virus (ISKNV) infection in aquarium-maintained mandarin fish, Siniperca chuatsi (Basilewsky), in Japan. J. Fish. Dis. 2014, 37, 401–405. [Google Scholar] [CrossRef] [PubMed]
- Figueiredo, P.; Tavares, G.C.; Dorella, F.A.; Rosa, J.C.C.; Marcelino, S.A.C.; Pierezan, F.; Pereira, F.L. First report of infectious spleen and kidney necrosis virus infection in Nile tilapia in Brazil. Transbound Emerg. Dis. 2021, 1–8. [Google Scholar] [CrossRef]
- OIE. Chapter 2.3.7. Red Sea Bream Iridoviral Diseases. 2019. Available online: https://www.oie.int/fileadmin/Home/eng/Internationa_Standard_Setting/docs/pdf/2.3.07_RSIVD.pdf (accessed on 20 June 2021).
- Jun, L.J.; Jeong, J.B.; Kim, J.H.; Nam, J.H.; Shin, K.W.; Kim, J.K.; Kang, J.; Jeong, H.D. Influence of temperature shifts on the onset and development of red sea bream iridoviral disease in rock bream Oplegnathus fasciatus. Dis. Aquat. Organ. 2009, 84, 201–208. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, J.G.; Zeng, K.; Weng, S.P.; Chan, S.M. Experimental transmission, pathogenicity and physical-chemical properities of infectious spleen and kidney necrosis virus (ISKNV). Aquaculture 2002, 204, 11–24. [Google Scholar] [CrossRef]
- Do, J.W.; Cha, S.J.; Kim, J.S.; An, E.J.; Lee, N.S.; Choi, H.J.; Lee, C.H.; Park, M.S.; Kim, J.W.; Kim, Y.C.; et al. Phylogenetic analysis of the major capsid protein gene of iridovirus isolates from cultured flounders Paralichthys olivaceus in Korea. Dis. Aquat. Organ. 2005, 64, 193–200. [Google Scholar] [CrossRef]
- Go, J.; Whittington, R. Experimental transmission, and virulence of a Megalocytivirus (Family Iridoviridae) of dwarf gourami (Colisa lalia) from Asia in Murray cod (Maccullochella peelii peelii) in Australia. Aquaculture 2006, 258, 140–149. [Google Scholar] [CrossRef]
- Weber, E.S.; Waltzek, T.B.; Young, D.A.; Twitchell, E.L.; Gates, A.E.; Vagelli, A.; Risatti, G.R.; Hedrick, R.P.; Frasca, S., Jr. Systemic iridovirus infection in the Banggai cardinalfish (Pterapogon kauderni Koumans, 1933). J. Vet. Diagn. Investig. 2009, 21, 306–320. [Google Scholar] [CrossRef] [PubMed]
- Weisburg, W.G.; Barns, S.M.; Pelletier, D.A.; Lane, D.J. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 1991, 173, 697–703. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gardell, A.M.; Qin, Q.; Rice, R.H.; Li, J.; Kültz, D. Derivation and osmotolerance characterization of three immortalized tilapia (Oreochromis mossambicus) cell lines. PLoS ONE 2014, 9, e95919. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Waiyamitra, P.; Tattiyapong, P.; Sirikanchana, K.; Mongkolsuk, S.; Nicholson, P.; Surachetpong, W. A TaqMan RT-qPCR assay for tilapia lake virus (TiLV) detection in tilapia. Aquaculture 2018, 497, 184–188. [Google Scholar] [CrossRef]
- Hanson, L.A.; Rudis, M.R.; Vasquez-Lee, M.; Montogomery, R.D. A broadly applicable method to characterize large DNA viruses and adenoviruses based on the DNA polymerase gene. Virol. J. 2006, 3, 28. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soto, E.; Bowles, K.; Fernandez, D.; Hawke, J.P. Development of a real-time PCR assay for identification and quantification of the fish pathogen Francisella noatunensis subsp. orientalis. Dis. Aquat. Organ. 2010, 89, 199–207. [Google Scholar] [CrossRef] [PubMed]
- Capella-Gutiérrez, S.; Silla-Martínez, J.M.; Gabaldón, T. trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 2009, 25, 1972–1973. [Google Scholar] [CrossRef] [PubMed]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; Haeseler, A.V.; Lanfear, R. IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Senapin, S.; Dong, H.T.; Meemetta, W.; Gannonngiw, W.; Sangsuriya, P.; Vanichviriyakit, R.; Sonthi, M.; Nuangsaeng, B. Mortality from scale drop disease in farmed Lates calcarifer in Southeast Asia. J. Fish. Dis. 2018, 42, 119–127. [Google Scholar] [CrossRef] [Green Version]
- de Groof, A.; Guelen, L.; Deijs, M.; van der Wal, Y.; Miyata, M.; Ng, K.S.; van Grinsven, L.; Simmelink, B.; Biermann, Y.; Grisez, L.; et al. A novel virus Causes Scale Drop Disease in Lates calcarifer. PLoS Pathog. 2015, 11, e1005074. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soto, E.; Lopez-Porras, A.; Kenelty, K.; Yun, S.; Pierezan, F. Analysis of the Nile tilapia (Oreochromis niloticus) immune response during Francisella noatunensis subsp. orientalis infection. In Proceedings of the 2019 AFS Fish Health Section Annual Meeting and 60th Western Fish Disease Workshop, Ogden, UT, USA, 17–20 June 2019. [Google Scholar]
- Siwicki, A.; Pozet, F.; Morand, M.; Terech-Majewska, E.; Bernard, D. Pathogenesis of iridovirus: In vitro influence on macrophage activity and cytokine-like protein production in fish. Acta Vet. Brno 2001, 70, 451–456. [Google Scholar] [CrossRef] [Green Version]
- Ni, S.; Yan, Y.; Cui, H.; Yu, Y.; Huang, Y.; Qin, Q. Fish miR-146a promotes Singapore grouper iridovirus infection by regulating cell apoptosis and NF-κB activation. J. Gen. Virol. 2017, 98, 1489–1499. [Google Scholar] [CrossRef] [PubMed]
- Gibson- Kueh, S.; Chee, D.; Chen, J.; Wang, Y.H.; Tay, S.; Leong, L.N.; Ng, M.L.; Jones, J.B.; Nicholls, P.; Ferguson, H.W. The pathology of ‘scale drop syndrome’ in Asian seabass, Lates calcarifer Bloch, a first description. J. Fish Dis. 2012, 35, 19–27. [Google Scholar] [CrossRef] [PubMed]
- McGorgan, D.G.; Ostland, V.E.; Bryne, P.J.; Ferguson, H.W. Systemic disease involving an iridovirus-like agent in cultured tilapia, Oreochromis niloticus L. A case report. J. Fish. Dis. 1998, 21, 149–152. [Google Scholar] [CrossRef] [PubMed]
- Wirth, W.; Schwarzkopf, L.; Skerratt, L.F.; Ariel, E. Ranaviruses and reptiles. PeerJ 2018, 6, e6083. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Williams, T.; Barbosa-Solomieu, V.; Chinchar, V.G. A decade of advances in iridovirus research. In Advances in Virus Research; Maramorosch, K., Shatkin, A.J., Eds.; Elsevier Academic Press Inc.: San Diego, CA, USA, 2005; Volume 65, p. 173. [Google Scholar]
- Jancovich, J.K.; Chinchar, V.G.; Hyatt, A.; Miyazaki, T.; Williams, T.; Zhang, Q.Y. Family Iridoviridae. In Virus Taxonomy: Ninth Report of the International Committee on Taxonomy of Viruses; King, A.M.Q., Adams, M.J., Carstens, E.B., Lefkowitz, E.J., Eds.; Elsevier Academic Press: San Diego, CA, USA, 2012; pp. 193–210. [Google Scholar]
- Huang, Y.; Cai, S.; Jian, J.; Liu, G.; Xu, L. Co-infection of infectious spleen and kidney necrosis virus and Francisella sp. in farmed pearl gentian grouper (♀Epinephelus fuscoguttatus ×♂E. lanceolatus) in China—A case report. Aquaculture 2020, 526, 735409. [Google Scholar] [CrossRef]
- Liu, X.; Sun, W.; Zhang, Y.; Zhou, Y.; Xu, J.; Gao, X.; Zhang, S.; Zhang, X. Impact of Aeromonas hydrophila and infectious spleen and kidney necrosis virus infections on susceptibility and host immune response in Chinese perch (Siniperca chuatsi). Fish Shellfish Immunol. 2020, 105, 117–125. [Google Scholar] [CrossRef] [PubMed]
- Landsberg, J.H.; Kiryu, Y.; Tabuchi, M.; Waltzek, T.B.; Enge, K.M.; Reintjes-Tolen, S.; Preston, A.; Pessier, A.P. Co-infection by alveolate parasites and frog virus 3-like ranavirus during an amphibian larval mortality event in Florida, USA. Dis. Aquat. Organ. 2013, 105, 89–99. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| Target Gene | Primer Sequence (5′-3′) | Reference |
|---|---|---|
| 16S rRNA gene | F: AGAGTTTGATCATGGCTCAG R: GGTTACCTTGTTACGACTT | [23] |
| TilV-segment 3 | TiLV-93F: GGWGARGATGTTCGTGAAGG TiLV-93R: CTGCTTGAGTTGTGCTTCT TiLV-93Probe: FAM-CTCTACCAGCTAGTGCCCCA-IowaBlack | [24] |
| DNA polymerase gene | Cons lower: CCCGAATTCAGATCTCNGTRTCNCCRTA (N = A/C/G/T, R = A/G) HV: CGGAATTCTAGAYTTYGCNWSNYTNTAYCC (S = C/G, W = A/T) | [25] |
| Tilapia Megalocytivirus Major Capsid protein | nIridoV_F: TGGGTTTGTCGACATTGCAT nIridoV_R: GTCGCCGCCGTAAAGCT nIridoV_probe: FAM-TTA+C+GATGCAATGGA+GG+AG-BHQ-1 | This study |
| Francisella orientalis iglC | IglC_F:GGGCGTATCTAAGGATGGTATGAG IglC_R:AGCACAGCATACAGGCAAGCTA IglC_probe: FAM ATCTATTGATGGGCTCACAACTTCACAA-BHQ-1 | [26] |
| Fish no. | Length (cm) | Weight (g) | External Signs | Internal Signs | Francisella orientalis (Fo) Detection | TiLV qPCR [24] | Large DNA Virus PCR [25] |
|---|---|---|---|---|---|---|---|
| F1 | 6 | 8.53 | 1-Damaged fin 2-Dark skin 3-Pale gills 4-Ascites | 1-Hepatomegaly, splenomegaly and renalomegally. 2-Whitish nodules on spleen and kidney | 1-MTM: + Fo in head kidney and–in brain 2-16SrRNA PCR and sequencing: 99.6% ANI% to Fo | – | + (609 bp) |
| F2 | 5 | 4.05 | 1-Damaged fin 2-Pale gills 3-Ascites | 1-Splenomegaly and renalomegally. 2-Whitish nodules on spleen | 1-MTM: + Fo in brain and head kidney 2-16SrRNA PCR and sequencing: 100% ANI% to Fo | – | + (609 bp) |
| F3 | 4 | 1.32 | NGC | NGC | – | NA | NA |
| F4 | 7.5 | 6.4 | NGC | NGC | – | NA | NA |
| F5 | 8 | 6.4 | 1-Dark skin 2-Ascites | 1-Splenomegaly 2-Whitish nodules on spleen | 1-MTM: + Fo in brain and head kidney 2-16SrRNA PCR and Sequencing: 100% ANI% to Fo | – | + (609 bp) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shahin, K.; Subramaniam, K.; Camus, A.C.; Yazdi, Z.; Yun, S.; Koda, S.A.; Waltzek, T.B.; Pierezan, F.; Hu, R.; Soto, E. Isolation, Identification and Characterization of a Novel Megalocytivirus from Cultured Tilapia (Oreochromis spp.) from Southern California, USA. Animals 2021, 11, 3524. https://doi.org/10.3390/ani11123524
Shahin K, Subramaniam K, Camus AC, Yazdi Z, Yun S, Koda SA, Waltzek TB, Pierezan F, Hu R, Soto E. Isolation, Identification and Characterization of a Novel Megalocytivirus from Cultured Tilapia (Oreochromis spp.) from Southern California, USA. Animals. 2021; 11(12):3524. https://doi.org/10.3390/ani11123524
Chicago/Turabian StyleShahin, Khalid, Kuttichantran Subramaniam, Alvin C. Camus, Zeinab Yazdi, Susan Yun, Samantha A. Koda, Thomas B. Waltzek, Felipe Pierezan, Ruixue Hu, and Esteban Soto. 2021. "Isolation, Identification and Characterization of a Novel Megalocytivirus from Cultured Tilapia (Oreochromis spp.) from Southern California, USA" Animals 11, no. 12: 3524. https://doi.org/10.3390/ani11123524
APA StyleShahin, K., Subramaniam, K., Camus, A. C., Yazdi, Z., Yun, S., Koda, S. A., Waltzek, T. B., Pierezan, F., Hu, R., & Soto, E. (2021). Isolation, Identification and Characterization of a Novel Megalocytivirus from Cultured Tilapia (Oreochromis spp.) from Southern California, USA. Animals, 11(12), 3524. https://doi.org/10.3390/ani11123524






